Journal of Computer and Communications, 2019, 7, 65-71
http://www.scirp.org/journal/jcc
ISSN Online: 2327-5227
ISSN Print: 2327-5219
Comparison of Spatiotemporal Fusion Models
for Producing High Spatiotemporal Resolution
Normalized Difference Vegetation Index Time
Series Data Sets
Zhizhong Han1, Wenya Zhao2
1College of Computer Science and Technology, Chongqing University of Posts and Telecommunications, Chongqing, China
2Chongqing Aerospace Poly Technic, Chongqing, China
How to cite this paper: Han, Z.Z. and
Zhao, W.Y. (2019) Comparison of Spati-
otemporal Fusion Models for Producing
High Spatiotemporal Resolution Norma-
lized Difference Vegetation Index Time
Series Data Sets. Journal of Computer and
Communications, 7, 65-71.
https://doi.org/10.4236/jcc.2019.77007
Received: May 15, 2019
Accepted: July 7, 2019
Published: July 10, 2019
Abstract
It has a great significance to combine multi-source with different spatial res-
olution and temporal resolution to produce high spatiotemporal resolution
Normalized Difference Vegetation Index (NDVI) time series data sets. In this
study, four spatiotemporal fusion models were analyzed and compared with
each other. The models included the spatial and temporal adaptive reflectance
model (STARFM), the enhanced spatial and temporal adaptive reflectance fu-
sion model (ESTARFM), the flexible spatiotemporal data fusion model
(FSDAF), and a spatiotemporal vegetation index image fusion model
(STVIFM). The objective of is to: 1) compare four fusion models using Land-
sat-MODIS NDVI image from the Banan district, Chongqing Province; 2)
analyze the prediction accuracy quantitatively and visually. Results indicate
that STVIFM would be more suitable to produce NDVI time series data sets.
Keywords
Spatiotemporal Fusion, NDVI, Time Series, STVIFM
1. Introduction
The Normalized Difference Vegetation Index (NDVI) is a widely used vegeta-
tion index (VI) and provides a way of evaluating the biophysical or biochemical
information related to vegetation growth [1]. Long term NDVI time-series data-
sets have been widely used for monitoring ecosystem dynamics to understand
the responses of climate change [2] [3]. However, due to financial and technical
DOI: 10.4236/jcc.2019.77007 Jul. 10, 2019
65
Journal of Computer and Communications
�
Z. Z. Han, W. Y. Zhao
constraints, it is difficult to obtain NDVI data with both high spatial and high
temporal resolution on the same remote sensing instrument [4]. In addition,
long periods of cloud cover problems in some regions have aggravated this mat-
ter [5]. Thus, spatiotemporal fusion techniques which combine NDVI date from
multi-sensors with high spatial and temporal resolution is feasible solution to
acquire remote sensing time series for monitoring surface vegetations dynamics
[6] [7].
Up to now, several spatiotemporal fusion models have been proposed. Gao et
al. [8] proposed a spatial and temporal adaptive reflectance fusion model
(STARFM) to blend MODIS and Landsat image to produce a synthetic surface
reflectance product at 30 m spatial resolution. Based the STARFM, Zhu et al. [9]
developed an enhanced spatial and temporal adaptive reflectance fusion model
(ESTARFM), introducing conversion coefficient between pixels and improving
the prediction accuracy. Zhu et al. [10] proposed the flexible spatiotemporal data
fusion model (FSDAF) which performs better in predicting abrupt land cover
changes. Liao et al. [11] developed a spatiotemporal vegetation index image fu-
sion model (STVIFM) to generate NDVI time series images with high spatial
and temporal resolution in heterogeneous regions. In this study, we made a
comparation between STARFM, ESTARFM, FSDAF, and STVIFM methods,
tested by Landsat and MODIS data acquired in same site and quantitatively as-
sess the accuracy of predicted image generated from each fusion model.
2. Materials and Methods
2.1. Study site and Data Preparation
In this study, a selected study area is shown in Figure 1, which located in Banan
District (29˚34'10''N, 106˚57'35''E) in Chongqing Province to perform the com-
parison between the spatiotemporal fusion models. We select MODIS daily sur-
face reflectance image and Landsat-8 image acquired for these dates during this
period: April 28, 2015, August 02, 2015, and October 21, 2015. All images are
pre-processed and calculated as NDVI data. Scene subset is shown in Figure 2.
DOI: 10.4236/jcc.2019.77007
Figure 1. Location of the study area.
66
Journal of Computer and Communications
�
Z. Z. Han, W. Y. Zhao
Figure 2. Landsat NDVI (upper row) and MODIS NDVI (lower row) images. From left
to right, they were acquired from April 28, 2015, August 02, 2015, and October 21, 2015,
respectively.
2.2. Selected Spatiotemporal Fusion Models
2.2.1. STARFM
The STARFM is based on the moving window technology, which requires at
least a pair of high-resolution image and coarse-resolution image on the base
time and one coarse-resolution image on the predicted time. By introducing a
weigh function using spectral difference, temporal difference and spatial differ-
ence to determining the contribution of other pixels in the window to the central
pixel. And then a synthetic high Spatiotemporal image (F(t2)) is predicted with
the high- and coarse-resolution data through the proposed weight function. This
model can be written as in Equation (1).
F t
( 2)
=
∑
Wi F t M t
( 1)
( 2)
+
(
−
M t
( 1))
(1)
where, F(t1) and M(t1) denote the high-and coarse resolution date on the base
date, M(t2) is the coarse resolution date at the predicted date, and Wi is the
weight function.
2.2.2. ESTARFM
The ESTARFM needs at least two pairs of high-resolution image and coarse res-
olution image on the base time and one coarse-resolution image on the pre-
dicted time. Compared with STARFM, this method not only considers the spa-
tial and spectral similarity between pixels, but also introduces a conversion coef-
ficient, which is derived from the high-and coarse-resolution data during the
observation period using a linear regression. The final high-resolution predic-
tion is computed as in Equation (2).
F t
( 2)
=
∑
Wi Vi F t M t
* (
( 1)
( 2)
+
−
M t
( 1))
(2)
DOI: 10.4236/jcc.2019.77007
67
Journal of Computer and Communications
�
Z. Z. Han, W. Y. Zhao
where, F(t1) and M(t1) denote the high-and coarse resolution data on the base
date, M(t2) is the coarse resolution data at the predicted date, and Wi, Vi denote
the weight function and conversion coefficient respectively.
2.2.3. FSDAF
The FSDAF using one pair of high-resolution image and coarse-resolution im-
age on the base time and one coarse-resolution image on the predicted time, and
it also need to use land cover map. This model integrates STARFM, the linear
unmixing method [12] and the thin plate spline (TPS) interpolator that main-
tains the land cover change signals and local variability, which combined the
temporal prediction from the linear unmixing method with the spatial predic-
tion obtained by the TPS and distribute the residual to fine pixel to get the final
prediction. It can be written as Equation (3).
F t
( 2)
=
F t
( 1)
+
∆∑
F
*
Wi
(3)
where, F(t1), F(t2) denote the high-resolution image on the base time and pre-
dicted time respectively. F∆ is referred to the change between t1 and t2, which
computed by the linear unmixing method and TPS. And Wi is the weight func-
tion.
2.2.4. STVIFM
The STVIFM requires two pairs of high- and coarse-resolution images acquired
on the base time and one coarse-resolution on the predicted date. On the one
hand, this model links the mean NDVI change of high-resolution pixels to mean
NDVI change of coarse resolution pixels within a moving window. On the other
hand, it also considers the difference in NDVI change rates at different growing
stages. And the final prediction can be written as Equation (4).
* NDVI
NDVI( 2) NDVI( 1)
(4)
∆∑
Wi
=
t
t
+
where, NDVI(t2), NDVI(t1) are the high-resolution date on the prediction time
and base time respectively. ΔNDVI denote the change between t1 and t2, which
calculated by this model. And the Wi is the weight function.
2.3. Assessing Prediction Accuracy
The model’s prediction performance is quantitatively evaluated by representative
metrics. And the r and RMSE (root mean squared errors) are used to measure
the difference between the predicted image and actual image. The formulations
of these metrics are as follows:
r
=
∑
∑
(
N
j
1
=
N
j
1
=
(
x
j
−
x
j
−
2
x
)
x y
)(
∑
N
j
=
−
y
)
j
(
y
j
2
−
y
)
(5)
2
DOI: 10.4236/jcc.2019.77007
RMSE
=
−
y
j
2
)
∑
N
j
=
1(
x
j
N
(6)
68
Journal of Computer and Communications
�
Z. Z. Han, W. Y. Zhao
where N is the total number of pixels in the predicted image, xj and yj are the
values of the jth pixel in the predicted image and the actual image respectively.
And x , y represent the mean gray values of the predicted image and the ac-
tual image respectively.
3. Result and Discussion
3.1. Prediction Performance
We use the August 02 Landsat NDVI image as validation source and use April
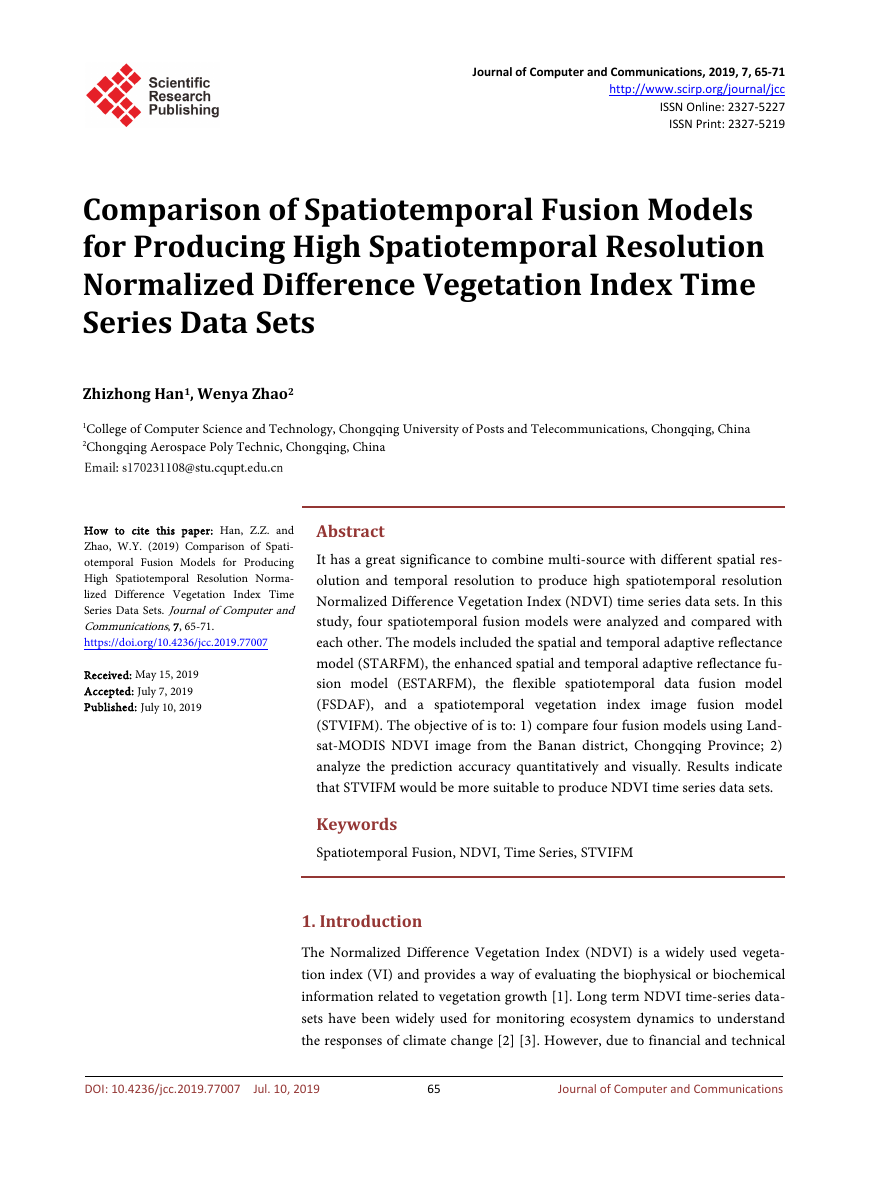
28 and October 21 to predict the August 02 image. Figure 3 shows the actual
NDVI image and predicted NDVI image by four spatiotemporal fusion models
on August 02, 2015. All the predicted NDVI images are consistent with the ac-
tual image from visual comparison, and water boundaries and clear land can be
predicted obviously, which demonstrate the practicality of these spatiotemporal
models.
3.2. Quantitative Assessment
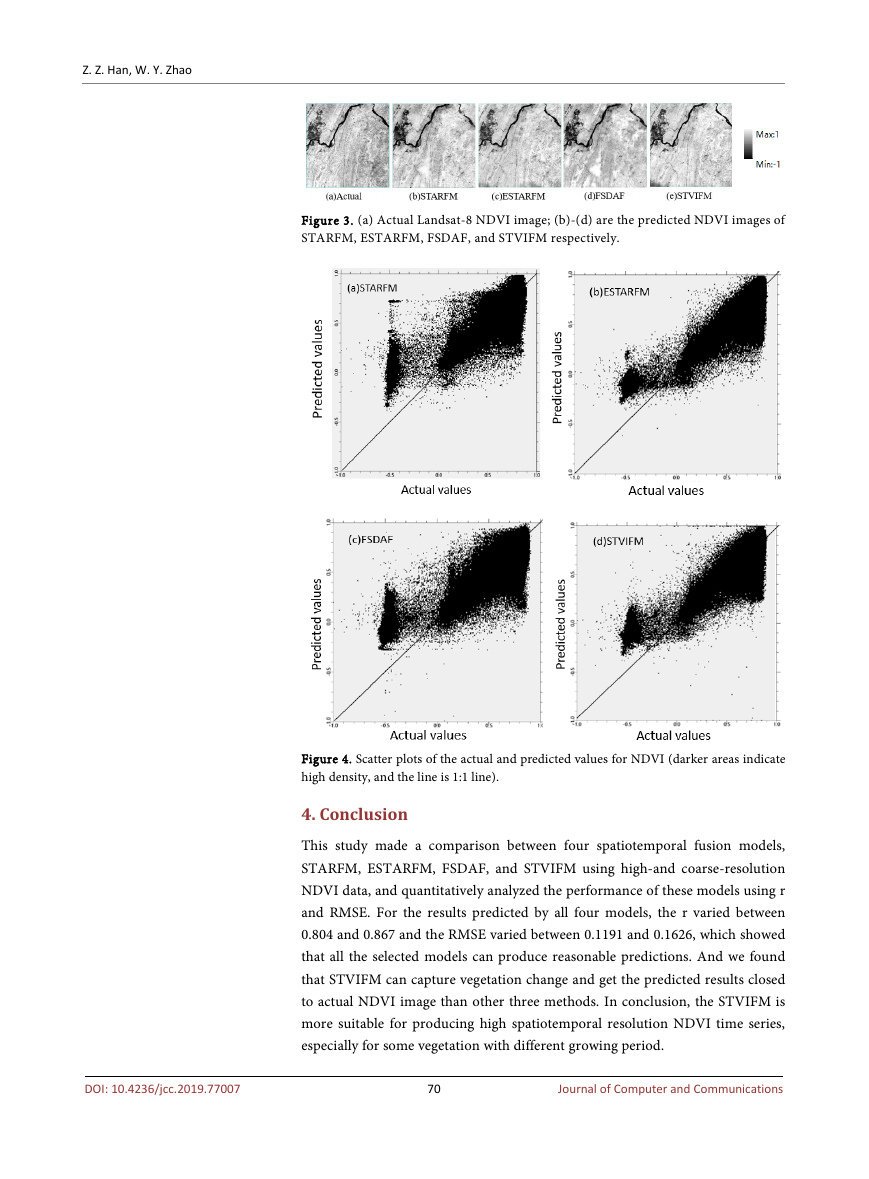
Scatter plots in Figure 4 indicate the difference between the actual NDVI values
and the predicted NDVI values on August 02 2015. We can see that the pre-
dicted NDVI values by four spatiotemporal fusion models are all fall close to the
1:1 line, which show all four spatiotemporal fusion models can capture changes
in phenology. And the prediction of ESTARFM and STVIFM using one input
pair is relatively accurate than that of STARFM and FSDAF using two input
pairs, which because two input pairs can provide more spatial details.
To better assess the accuracy of predictions, the metrics r and RMSE were
calculated in Table 1. All four methods can get the change details to the base
date image to get the prediction. The accuracy of the predicted NDVI image us-
ing the STVIFM is the best (r = 0.864, RMSE = 0.1191) and a little better than
the accuracy of the predicted NDVI image using ESTARFM (r = 0.867, RMSE =
0.1247). The image predicted by STARFM (r = 0.804, RMSE = 0.1626) and
FSDAF (r = 0.810, RMSE = 0.1446) can also produce an accurate result, but
these two models got inaccurate predictions on some pixels (Figure 3(b), Figure
3(d)), which demonstrate the predictions using two input pairs is relatively
more accurate.
Table 1. Comparison of rand RMSE betweeen actual NDVI and predicted NDVI by using
STARFM, ESTARFM, FSDAF, and STVIFMmodelsin the study area on August 02 2015.
Models
STARFM
ESTARFM
FSDAF
STVIFM
r
0.804
0.867
0.810
0.864
RMSE
0.1626
0.1247
0.1446
0.1191
69
Journal of Computer and Communications
DOI: 10.4236/jcc.2019.77007
�
Z. Z. Han, W. Y. Zhao
Figure 3. (a) Actual Landsat-8 NDVI image; (b)-(d) are the predicted NDVI images of
STARFM, ESTARFM, FSDAF, and STVIFM respectively.
Figure 4. Scatter plots of the actual and predicted values for NDVI (darker areas indicate
high density, and the line is 1:1 line).
4. Conclusion
This study made a comparison between four spatiotemporal fusion models,
STARFM, ESTARFM, FSDAF, and STVIFM using high-and coarse-resolution
NDVI data, and quantitatively analyzed the performance of these models using r
and RMSE. For the results predicted by all four models, the r varied between
0.804 and 0.867 and the RMSE varied between 0.1191 and 0.1626, which showed
that all the selected models can produce reasonable predictions. And we found
that STVIFM can capture vegetation change and get the predicted results closed
to actual NDVI image than other three methods. In conclusion, the STVIFM is
more suitable for producing high spatiotemporal resolution NDVI time series,
especially for some vegetation with different growing period.
70
Journal of Computer and Communications
DOI: 10.4236/jcc.2019.77007
�
Z. Z. Han, W. Y. Zhao
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this pa-
per.
References
[1] Busetto, L., Meroni, M. and Colombo, R. (2008) Combining Medium and Coarse
Spatial Resolution Satellite Data to Improve the Estimation of Sub-Pixel NDVI
Time Series. Remote Sens. Environ., 112, 118-131.
https://doi.org/10.1016/j.rse.2007.04.004
[2] Tewes, A., Thonfeld, F., Schmidt, M., Oomen, R., Zhu, X., Dubovyk, O., Menz, G.
and Schellberg, J. (2015) Using RapidEye and MODIS Data Fusion to Monitor Ve-
getation Dynamics in Semi-Arid Rangelands in South Africa. Remote Sens., 7,
6510-6534. https://doi.org/10.3390/rs70606510
[3] Bhandari, S., Phinn, S. and Gill, T. (2012) Preparing Landsat Image Time Series
(LITS) for Monitoring Changes in Vegetation Phenology in Queensland, Australia.
Remote Sens., 4, 1856-1886. https://doi.org/10.3390/rs4061856
[4] Gevaert, C.M. and García-Haro, F.J. (2015) A Comparison of STARFM and an
Unmixing Based Algorithm for Landsat and MODIS Data Fusion. Remote Sensing
of Environment, 156, 34-44. https://doi.org/10.1016/j.rse.2014.09.012
[5] Schmidt, M., Udelhoven, T., Gill, T. and Röder, A. (2012) Long Term Data Fusion
for a Dense Time Series Analysis with MODIS and Landsat Imagery in an Australi-
an Savanna. J. Appl. Remote Sens., 6, 63512. https://doi.org/10.1117/1.JRS.6.063512
[6] Fensholt, R. (2004) Earth Observation of Vegetation Status in the Sahelian and Su-
danian West Africa: Comparison of Terra MODIS and NOAA AVHRR Satellite
Data. Int. J. Remote Sens., 25, 1641-1659.
https://doi.org/10.1080/01431160310001598999
[7] Zurita-Milla, R., Clevers, J., van Gijsel, J. and Schaepman, M. (2011) Using MERIS
Fused Images for Classification Mapping and Vegetation Status Assessment in He-
terogeneous Landscapes. Int. J. Remote Sens., 32, 973-991.
https://doi.org/10.1080/01431160903505286
[8] Gao, F., Masek, J., Schwaller, M. and Hall, F. (2006) On the Blending of the MODIS
and Landsat ETM+ Surface Reflectance. IEEE Trans. Geosci. Remote Sens., 44,
2207-2218. https://doi.org/10.1109/TGRS.2006.872081
[9] Zhu, X., Chen, J., Gao, F., Chen, X. and Masek, J.G. (2010) An enhanced Spatial and
Temporal Adaptive Reflectance Fusion Model for Complex Heterogeneous Regions.
Remote Sens. Environ., 114, 2610-2623. https://doi.org/10.1016/j.rse.2010.05.032
[10] Zhu, X., Helmer, E.H., Gao, F., Liu, D., Chen, J. and Lefsky, M.A. (2016) A Flexible
Spatiotemporal Method for Fusing Satellite Images with Different Resolutions. Re-
mote Sens. Environ., 172, 165-177. https://doi.org/10.1016/j.rse.2015.11.016
[11] Liao, C., Wang, J., Pritchard, I., et al. (2017) A Spatio-Temporal Data Fusion Model
for Generating NDVI Time Series in Heterogeneous Regions. Remote Sensing, 9,
1125. https://doi.org/10.3390/rs9111125
[12] Zhukov, B., Oertel, D. and Lanzl, F. (1999) Unmixing-Based Multisensor Multire-
solution Image Fusion. IEEE Trans. Geosci. Remote Sens., 37, 1212-1226.
https://doi.org/10.1109/36.763276
71
Journal of Computer and Communications
DOI: 10.4236/jcc.2019.77007
�












 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc