Finding overlapping communities in networks by label
propagation
Steve Gregory
Department of Computer Science, University of Bristol, Bristol BS8 1UB, England
Abstract. We propose an algorithm for finding overlapping community structure in very large
networks. The algorithm is based on the label propagation technique of Raghavan, Albert, and
Kumara, but is able to detect communities that overlap. Like the original algorithm, vertices
have labels that propagate between neighbouring vertices so that members of a community
reach a consensus on their community membership. Our main contribution is to extend the
label and propagation step to include information about more than one community: each
vertex can now belong to up to v communities, where v is the parameter of the algorithm. Our
algorithm can also handle weighted and bipartite networks. Tests on an independently-
designed set of benchmarks, and on real networks, show the algorithm to be highly effective in
recovering overlapping communities. It is also very fast and can process very large and dense
networks in a short time.
1. Introduction
Networks are a natural representation of various kinds of complex system, in society, biology, and
other fields. Although the study of networks is not new, the amount of network data has proliferated
in recent years, thanks to developments in computing and communications technology. As the number
and size of network datasets has increased, so too has the interest in computational techniques that
help us to understand the properties of networks.
A key property of many networks is their community structure: the tendency for vertices to be
gathered into distinct groups, or communities, such that edges between vertices in the same
community are dense but intercommunity edges are sparse. Identifying communities can allow us to
understand attributes of vertices from the network topology alone. For example, all vertices in a
community may be somehow related, or a vertex that appears in more than one community may play
a special role. The automatic discovery of network communities can also help reveal the coarse-
grained structure of networks which are too large for humans to make sense of at the level of
individual vertices.
A vast number of community detection algorithms have been developed, especially in the last few
years. Most of them handle unipartite networks with undirected, unweighted edges, and they vary in
performance and speed. The algorithms use a wide variety of techniques, including removal of high-
betweenness edges [1], optimization of modularity [2], detection of dense subgraphs [3], statistical
inference [4], and many more. Even a cursory description of these algorithms is well beyond the
scope of this paper. The interested reader is referred to Fortunato’s excellent, comprehensive survey
[5] of community detection.
The design of community detection algorithms seems to require a precise definition of community,
but unfortunately no such definition exists [5, 6]. Each algorithm makes different assumptions
consistent with the intuitive concept of a community. The majority of algorithms assume that a
network contains a flat set of disjoint communities. This makes sense for many networks: for
example, most employees work for a single employer, most papers are published in a single
�
conference, etc. Some algorithms [3, 7–10, 13] allow communities to overlap, each vertex possibly
appearing in more than one community. This is sometimes more realistic: for example, many
researchers belong to more than one research community. Other algorithms [11–13] can find a
hierarchy of communities: for example, a number of research communities each subdivided into
research groups.
As network datasets become larger, the speed of community detection algorithms becomes more
important. In future, algorithms will need to handle networks with millions of vertices in a reasonable
time, and so any practical algorithm must have a very low time complexity.
One of the fastest algorithms proposed to date is the label propagation algorithm of Raghavan et al
[14], which we shall call the RAK algorithm. As well as its near-linear time complexity (for sparse
networks), it is very simple and has no parameters. However, like most community detection
algorithms, it can detect only disjoint communities. In this paper, we propose an algorithm that
generalizes the RAK algorithm to find overlapping communities. It takes a parameter, v, which
controls the potential degree of overlap between communities. The RAK algorithm is essentially a
special case of the proposed algorithm with v=1.
The next section describes the RAK algorithm. Section 3 explains and justifies the design of our
algorithm, named COPRA (Community Overlap PRopagation Algorithm). In section 4 we present
the results of experiments that show how our algorithm behaves, and measure its performance and
speed, comparing these with other overlapping community detection algorithms. Section 5 shows how
the algorithm can be simply extended to handle bipartite networks. Conclusions appear in section 6.
2. Detecting communities by label propagation
The RAK algorithm can be described very simply. Each vertex is associated with a label, which is an
identifier such as an integer.
1. To initialize, every vertex is given a unique label.
2. Then, repeatedly, each vertex x updates its label by replacing it by the label used by the
greatest number of neighbours. If more than one label is used by the same maximum
number of neighbours, one of them is chosen randomly. After several iterations, the same
label tends to become associated with all members of a community.
3. All vertices with the same label are added to one community.
The propagation phase does not always converge to a state in which all vertices have the same
label in successive iterations. To ensure that the propagation phase terminates, Raghavan et al propose
the use of “asynchronous” updating, whereby vertex labels are updated according to the previous label
of some neighbours and the updated label of others. Vertices are placed in some random order. x’s
new label in the tth iteration is based on the labels of the neighbours that precede x in the tth iteration
and the labels of its neighbours that follow x in the (t-1)th iteration.
The algorithm terminates when every vertex has a label that is one of those that are used by a
maximum number of neighbours.
The algorithm produces groups that contain all vertices sharing the same label. These groups are
not necessarily connected, in the sense that there is a path between every pair of vertices in the group
passing only through vertices in the same group. Since communities are generally required to be
connected, Raghavan et al propose a final phase that splits the groups into one or more connected
communities.
The time complexity of the algorithm is almost linear in the network size. Initialization takes time
O(n), each iteration takes time O(m), and the time for processing disconnected communities is
O(m+n). The number of iterations required is harder to predict, but Raghavan et al claim that five
iterations is sufficient to classify 95% of vertices correctly.
Leung et al [15] have analysed the RAK algorithm in more detail. They compare asynchronous
with synchronous updating, whereby the new label of each vertex in the ith iteration is always based
on the labels of its neighbours in the (i-1)th iteration. They found that synchronous updating requires
more iterations than asynchronous updating, but is “much more stable”. They also propose restraining
the propagation of labels to limit the size of communities, and a similar technique to allow detection
of hierarchical communities. Both Refs. [14] and [15] hint at the possibility of detecting overlapping
communities, but neither extends the algorithm to find them; we do this in the next section.
�
3. Overlapping communities
3.1. Extending to overlapping communities
In the RAK algorithm, a vertex label identifies a single community to which the vertex belongs. If
communities overlap, each vertex may belong to more than one community. Therefore, to find
overlapping communities, we clearly need to allow a vertex label to contain more than one
community identifier.
As a first attempt, we might let a vertex label be a set of community identifiers. Initially, each
vertex would be labelled by a unique identifier, and each propagation step would copy all of x’s
neighbours’ community identifiers into x’s set. This clearly will not work, because every vertex would
end up labelled with the set of all community identifiers.
Alternatively, we could label each vertex x with a set of pairs (c,b), where c is a community
identifier and b is a belonging coefficient, indicating the strength of x’s membership of community c,
such that all belonging coefficients for x sum to 1. Each propagation step would set x’s label to the
union of its neighbours’ labels, sum the belonging coefficients of the communities over all
neighbours, and normalize. More precisely, assuming a function bt(c,x) that maps a vertex x and
community identifier c to its belonging coefficient in iteration t,
(
xcb
t
,
)
=
,(
yc
1
b
∑
t
)(
xNy
)(
xN
)
,
(1)
where N(x) denotes the set of neighbours of x. We use synchronous updating, partly because it seems
to give better results than asynchronous updating [15]: in our algorithm, the label of a vertex in
iteration t is always based on its neighbours’ labels in iteration t-1.
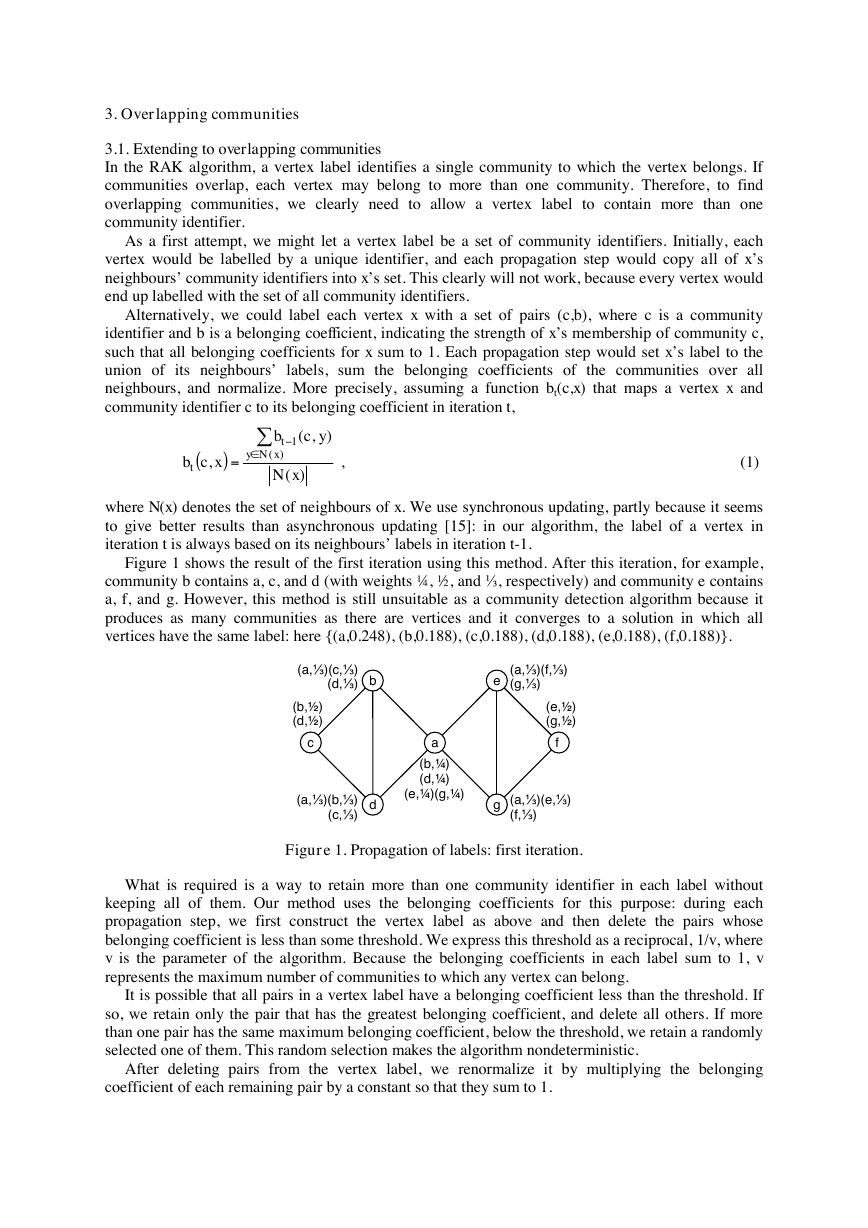
Figure 1 shows the result of the first iteration using this method. After this iteration, for example,
community b contains a, c, and d (with weights ¼, ½, and ⅓, respectively) and community e contains
a, f, and g. However, this method is still unsuitable as a community detection algorithm because it
produces as many communities as there are vertices and it converges to a solution in which all
vertices have the same label: here {(a,0.248), (b,0.188), (c,0.188), (d,0.188), (e,0.188), (f,0.188)}.
(a,⅓)(c,⅓)
(d,⅓)
b
(a,⅓)(f,⅓)
(g,⅓)
e
(b,½)
(d,½)
c
(a,⅓)(b,⅓)
(c,⅓)
d
a
(b,¼)
(d,¼)
(e,¼)(g,¼)
(e,½)
(g,½)
f
g
(a,⅓)(e,⅓)
(f,⅓)
Figure 1. Propagation of labels: first iteration.
What is required is a way to retain more than one community identifier in each label without
keeping all of them. Our method uses the belonging coefficients for this purpose: during each
propagation step, we first construct the vertex label as above and then delete the pairs whose
belonging coefficient is less than some threshold. We express this threshold as a reciprocal, 1/v, where
v is the parameter of the algorithm. Because the belonging coefficients in each label sum to 1, v
represents the maximum number of communities to which any vertex can belong.
It is possible that all pairs in a vertex label have a belonging coefficient less than the threshold. If
so, we retain only the pair that has the greatest belonging coefficient, and delete all others. If more
than one pair has the same maximum belonging coefficient, below the threshold, we retain a randomly
selected one of them. This random selection makes the algorithm nondeterministic.
After deleting pairs from the vertex label, we renormalize it by multiplying the belonging
coefficient of each remaining pair by a constant so that they sum to 1.
˛
-
�
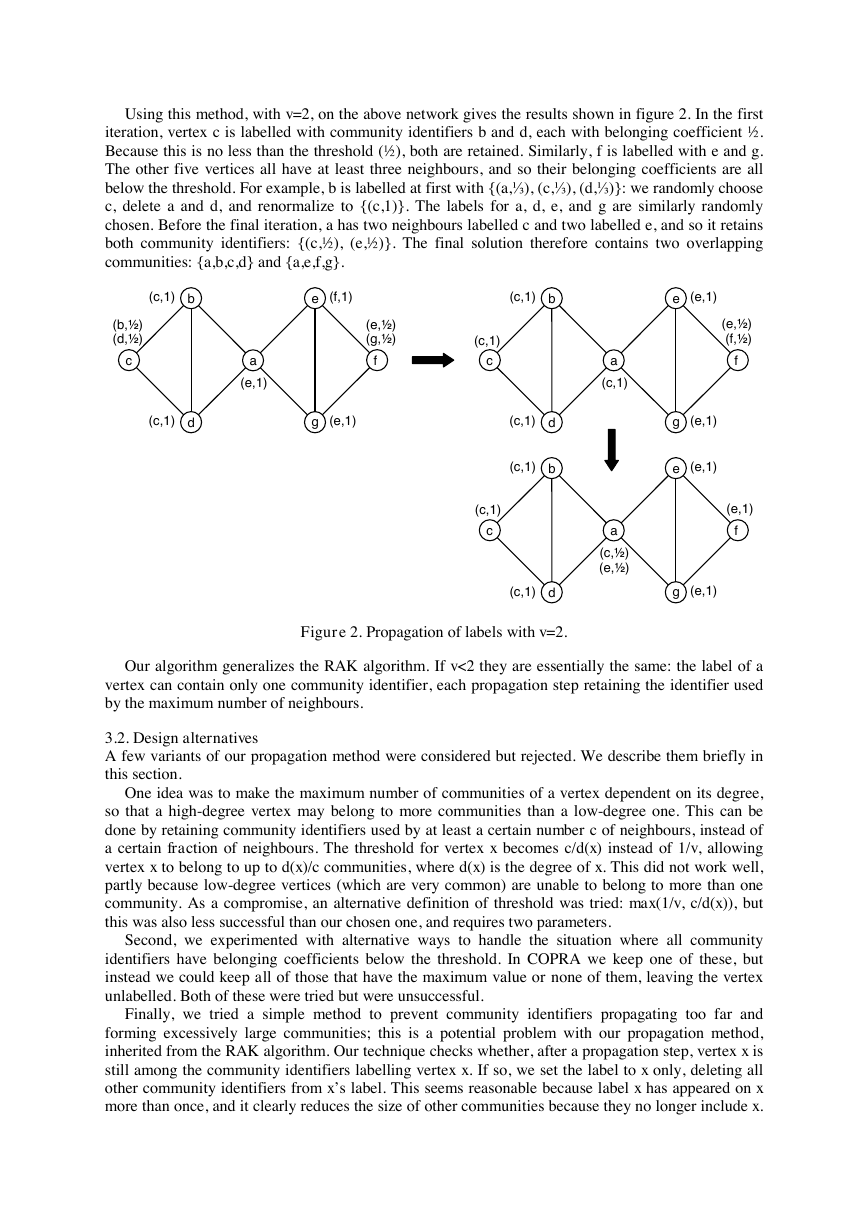
Using this method, with v=2, on the above network gives the results shown in figure 2. In the first
iteration, vertex c is labelled with community identifiers b and d, each with belonging coefficient ½.
Because this is no less than the threshold (½), both are retained. Similarly, f is labelled with e and g.
The other five vertices all have at least three neighbours, and so their belonging coefficients are all
below the threshold. For example, b is labelled at first with {(a,⅓), (c,⅓), (d,⅓)}: we randomly choose
c, delete a and d, and renormalize to {(c,1)}. The labels for a, d, e, and g are similarly randomly
chosen. Before the final iteration, a has two neighbours labelled c and two labelled e, and so it retains
both community identifiers: {(c,½), (e,½)}. The final solution therefore contains two overlapping
communities: {a,b,c,d} and {a,e,f,g}.
(c,1)
b
e
(f,1)
(c,1)
b
e
(e,1)
(b,½)
(d,½)
c
a
(e,1)
(e,½)
(g,½)
f
(c,1)
d
g
(e,1)
(c,1)
c
(c,1)
c
d
(c,1)
(c,1)
b
a
(c,1)
a
(c,½)
(e,½)
(e,½)
(f,½)
f
(e,1)
f
g
(e,1)
e
(e,1)
Figure 2. Propagation of labels with v=2.
(c,1)
d
g
(e,1)
Our algorithm generalizes the RAK algorithm. If v<2 they are essentially the same: the label of a
vertex can contain only one community identifier, each propagation step retaining the identifier used
by the maximum number of neighbours.
3.2. Design alternatives
A few variants of our propagation method were considered but rejected. We describe them briefly in
this section.
One idea was to make the maximum number of communities of a vertex dependent on its degree,
so that a high-degree vertex may belong to more communities than a low-degree one. This can be
done by retaining community identifiers used by at least a certain number c of neighbours, instead of
a certain fraction of neighbours. The threshold for vertex x becomes c/d(x) instead of 1/v, allowing
vertex x to belong to up to d(x)/c communities, where d(x) is the degree of x. This did not work well,
partly because low-degree vertices (which are very common) are unable to belong to more than one
community. As a compromise, an alternative definition of threshold was tried: max(1/v, c/d(x)), but
this was also less successful than our chosen one, and requires two parameters.
Second, we experimented with alternative ways to handle the situation where all community
identifiers have belonging coefficients below the threshold. In COPRA we keep one of these, but
instead we could keep all of those that have the maximum value or none of them, leaving the vertex
unlabelled. Both of these were tried but were unsuccessful.
Finally, we tried a simple method to prevent community identifiers propagating too far and
forming excessively large communities; this is a potential problem with our propagation method,
inherited from the RAK algorithm. Our technique checks whether, after a propagation step, vertex x is
still among the community identifiers labelling vertex x. If so, we set the label to x only, deleting all
other community identifiers from x’s label. This seems reasonable because label x has appeared on x
more than once, and it clearly reduces the size of other communities because they no longer include x.
�
This technique was successful in improving the worst results but the best results deteriorated,
probably because community sizes were sometimes limited more than they should have been. Leung
et al. [15] proposed a more complicated “hop attenuation” strategy to achieve a similar effect, also
with inconclusive results.
3.3. Termination
Like the RAK algorithm, our algorithm often does not converge to a state in which vertex labels
remain constant between iterations. Community identifiers may continue to propagate indefinitely (for
example, around a four-cycle). Ref. [14] solves this by a termination criterion that is unsuitable for
COPRA because our vertex labels are more complex and we use synchronous updating. We therefore
need a new termination condition.
In our algorithm, the number of community identifiers is initially n (the number of vertices) and
decreases monotonically as identifiers are deleted from the vertex labels. Eventually this number
reaches a minimum, even if this is 1 (a trivial solution). That is, the set of community identifiers in
use at the tth iteration,
=
i
t
{
xcbVxVc
)}0),(
(
:
,
(2)
>
t
where V is the set of all vertices, reaches a minimum size.
The propagation should not terminate as soon as it = it-1, unless |it| = 1, because it may reduce again
after many more propagation steps. Even if it does not, the solution might improve further as
community identifiers continue to move between vertices. Instead, we could check the number of
vertices labelled with each community identifier,
=
c
t
:),{(
ic
iVc
=
}
1
∑
>
0),(
xcbVx
,
t
,
(3)
but this usually changes in successive iterations, so we cannot expect that ct = ct-1.
Our chosen method is to compute the minimum number of vertices labelled with each community
identifier since the number of identifiers last reduced:
=
,
)
:),{(
ic
pcqp
((
mt =
mt = ct otherwise
(4)
We stop the propagation as soon as mt = mt-1. It is easy to see that this will happen after a finite
number of steps, and so the algorithm is guaranteed to terminate.
if it = it-1
),(
qc
min(
c
t
,
qp
))}
c
t
1
i
Although we cannot prove that the tth iteration always represents the best solution, our termination
criterion works well in practice and is inexpensive to compute. We use it for all results presented in
section 4.
3.4. Postprocessing
After termination, each vertex whose label contains community identifier c is simply allocated to
community c. However, this method might form communities that are subsets of (or identical to)
others. We avoid this by keeping track of the subset relationship between communities as they are
constructed, and then deleting any community that is a subset of any other. This is faster than
checking for set inclusion after all communities are constructed: it can be done in linear time.
As with the RAK algorithm, the communities found may not be connected, and so finally we split
all disconnected communities into smaller, connected ones.
3.5. The COPRA algorithm
The complete COPRA algorithm is shown in figures 3 and 4. It keeps two vectors of vertex labels: old
and new; old.x (resp. new.x) denotes the previous (resp. updated) label for vertex x. Each vertex label
is a set of pairs (c,b), where c is a community identifier and b is the belonging coefficient. N(x) is the
set of neighbours of vertex x.
3.6. Complexity
The time complexity of each step of the algorithm is estimated below. n is the number of vertices, m is
the number of edges, and v is the parameter (maximum number of communities per vertex).
˛
$
˛
˛
˛
˛
˛
$
$
-
�
1. Initialization takes time O(n).
2. This phase constructs an updated label (maximum size O(vm/n)) for each of the n vertices.
For each vertex x, it iterates through the O(m/n) neighbours. For each neighbour y, it
iterates through all (at most v) community identifiers in y’s label, looking up each one in
x’s updated label (which takes time O(log (vm/n)) if a tree data structure is used). The
normalization and deletion activities for each vertex x iterate through the updated label of x
(time O(vm/n) per vertex). The time for the whole phase is therefore O(vm log (vm/n)).
3. This iterates through all O(v) community identifiers of each of the n vertices, looking up
each in a set of size O(n). Using a hash table data structure, the total time is O(vn).
4. Similar to phase 3. The total time is O(vn).
5. Iterates through all community identifiers, c, of all vertices, x (O(vn) steps). At each step, it
adds x to community c and updates the set of communities of which c is a subset (time
O(v2), because the set sizes are at most v). The total time is O(v3n).
6. Iterates through at most n communities: time O(n).
7. This is similar to the RAK algorithm but there are up to vn, instead of n, vertices in the
communities at this stage, and so the total time is O(v(m+n)).
Phases 2-4 are repeated, so the time per iteration is O(vm log (vm/n)). The initial and final steps
take time O(v(m+n)+v3n).
For a sparse network, the time complexity is therefore O(v3n) plus O(vn log (v)) per iteration.
Assuming that v is a small integer the execution time per iteration is essentially linear.
3.7. Weighted networks
To extend COPRA to weighted networks, we simply replace the line b ‹
by, in the Propagate
operation, by b ‹
bywxy, where wxy is the weight of the edge {x,y}.
1. For each vertex x:
old.x ‹
{(x,1)}.
2. For each vertex x:
Propagate(x, old, new).
3. If id(old) = id(new):
mc(min, count(new)).
min ‹
Else:
min ‹
4. If min „
old ‹
oldmin ‹
count(new).
oldmin:
new.
Repeat from step 2.
min.
ids ‹
5. For each vertex x:
id(old.x).
For each c in ids:
coms ‹
sub ‹
Else:
coms ¨
sub ¨
6. For each (c,i) in sub:
coms ‹
sub ‹
If i „
{}: coms ‹
If, for some g, (c,g) is in coms, (c,i) in sub:
{(c,g¨ {x})}.
coms – {(c,g)} ¨
sub – {(c,i)} ¨
{(c,i˙
ids)}.
{(c,{x})}.
{(c,ids)}.
coms – (c,g).
7. Split disconnected communities in coms.
Figure 3. The COPRA algorithm.
�
{(c,bx+b)}.
{(c,b)}.
Propagate(x, source, dest):
by.
b ‹
If, for some bx, (c,bx) is in dest.x:
dest.x – {(c,bx)} ¨
dest.x ‹
{}.
dest.x ¨
dest.x ‹
For each y in N(x):
For each (c,by) in source.y:
Else: dest.x ‹
Normalize(dest.x).
bmax = 0.
For each (c,b) in dest.x:
b.
c.
If dest.x = {}: dest.x ‹
Else: Normalize(dest.x).
If b < 1/v:
dest.x ‹
If b > bmax:
bmax ‹
cmax ‹
dest.x – {(c,b)}.
{(cmax,1)}.
Normalize(l):
0.
sum ‹
For each (c,b) in l: sum ‹
For each (c,b) in l: l ‹
sum + b.
l – {(c,b)} ¨
{(c,b/sum)}.
{}.
id(l):
ids ‹
For each l.x in l: ids ‹
Return ids.
ids ¨
id(l.x).
{}.
id(x):
ids ‹
For each (c,b) in x: ids ‹
Return ids.
ids ¨
{c}.
{}.
count(l):
counts ‹
For each l.x in l:
For each (c,b) in l.x:
Return counts.
Else: counts ‹
counts ‹
If, for some n, (c,n) is in counts:
counts – {(c,n)} ¨
{(c,n+1)}.
counts ¨
{(c,1)}.
mc(cs1, cs2):
{}.
cs ‹
For each (c,n1) in cs1, (c,n2) in cs2:
cs ¨
Return cs.
{(c, min(n1,n2))}.
cs ‹
Figure 4. The COPRA algorithm (contd.).
�
4. Experiments
4.1. Methodology
There are two ways to evaluate the performance of a community detection algorithm. One is to run
the algorithm on real-world network data. A problem with this is that it is hard to judge the
communities found because we usually do not know the real communities that are present in the
original data; even if we do, they are not necessarily reflected in the network structure. The other
method is to use randomly-generated synthetic networks based on a known community structure and
compare the known communities with those found by the algorithm. A benefit of this method is that
we can vary the parameters of the networks to analyse the algorithm’s behaviour in detail. The
drawback is that artificial networks might not share the properties of real networks.
For synthetic networks, there are various standard measures [16–18] that can be used to compare
the known and computed communities, but none of these are suitable for overlapping communities.
Exceptions are the Omega index [19], which is based on the Adjusted Rand index, and a new variant
of the Mutual Information measure [13], extended to handle overlapping communities. We use the
Normalized Mutual Information (NMI) measure of Ref. [13] in the experiments reported in this paper.
For real networks, the quality of a solution is usually assessed by the relative density of edges
within communities and between communities. The most common measure of this is modularity [20,
21]: a high value indicates more intracommunity edges than would be expected by chance. The
original modularity measure is defined only for disjoint communities, but Nicosia et al [22] have
recently designed a variant that is defined also for overlapping communities. We use this overlap
modularity (Qov) measure for the experiments in this paper. Its value depends on the number of
communities to which each vertex belongs and the strength of its membership to each community; we
assume that each vertex belongs equally to all of the communities of which it is a member.
The overlap modularity measure is defined in terms of a function f, which in turn is defined as:
)(
xf
= 2
px
p
,
(5)
but the value of p is not specified in Ref. [22]. Therefore, we use the following definition of f,
suggested in Ref. [23]:
=
)(
xf
60
x
(6)
30
.
As well as measures of solution quality, we measure:
1. The number of non-singleton communities. This is more reliable than the total number of
communities because some algorithms assign every vertex to at least one community even
if it has only one member, while others leave some vertices unassigned to any community.
2. The overlap: the average number of communities to which each vertex belongs. This is the
sum of the sizes of all communities (including singletons) divided by the number of
vertices, n.
3. The execution time. This was measured using the programs running under Linux on an
AMD Opteron 250 CPU at 2.4GHz.
This section describes experiments on both artificial and real networks. For the former, we use the
network generator of Lancichinetti et al [25]. This produces benchmark networks which are claimed
to possess properties found in real networks, such as heterogeneous distributions of degree and
community size. Although not described in Ref. [25], the generator also allows communities to
overlap, with the restriction that every overlapping vertex belongs to a fixed number of communities.
The parameters of the benchmark networks are: the number of vertices (N), the average degree
(〈k〉), the mixing parameter (µ – each vertex shares a fraction µ of its edges with vertices in other
communities), the minimum community size (cmin), and the number of vertices that are in more than
one community (on).
Like the experiments reported in Ref. [26], we keep the remaining parameters fixed for all of our
experiments: the maximum degree kmax is 2.5*〈k〉, the maximum community size cmax is 5*cmin, and
each overlapping vertex belongs to two communities (om=2). The exponents of the power-law
distribution of vertex degrees (τ1) and community sizes (τ2) are -2 and -1, respectively.
-
-
�














 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc