REVIEW
Computer-aided diagnostic models in
breast cancer screening
Mammography is the most common modality for breast cancer detection and diagnosis and is often
complemented by ultrasound and MRI. However, similarities between early signs of breast cancer and
normal structures in these images make detection and diagnosis of breast cancer a difficult task. To aid
physicians in detection and diagnosis, computer-aided detection and computer-aided diagnostic (CADx)
models have been proposed. A large number of studies have been published for both computer-aided
detection and CADx models in the last 20 years. The purpose of this article is to provide a comprehensive
survey of the CADx models that have been proposed to aid in mammography, ultrasound and MRI
interpretation. We summarize the noteworthy studies according to the screening modality they consider
and describe the type of computer model, input data size, feature selection method, input feature type,
reference standard and performance measures for each study. We also list the limitations of the existing
CADx models and provide several possible future research directions.
Turgay Ayer1,
Mehmet US Ayvaci1,
Ze Xiu Liu1,
Oguzhan Alagoz1,2
& Elizabeth S
Burnside†1,3
1Industrial & Systems Engineering
Department, University of Wisconsin,
Madison, WI, USA
2Department of Population Health
Sciences, University of Wisconsin,
Madison, WI, USA
3Department of Biostatistics & Medical
Informatics, University of Wisconsin,
Madison, WI, USA
†Author for correspondence:
Department of Radiology, University
of Wisconsin Medical School, E3/311,
600 Highland Avenue, Madison,
WI 53792-3252, USA
Tel.: +1 608 265 2021
Fax: +1 608 265 1836
eburnside@uwhealth.org
KEYWORDS: breast cancer n computer-aided detection n computer-aided diagnosis
n mammography n MRIn ultrasound
Radiological imaging, which often includes
mammography, ultrasound (US) and MRI,
is the most effective means, to date, for early
detection of breast cancer [1]. However, differen-
tiating between benign and malignant findings
is difficult.
Successful breast cancer diagnosis requires sys-
tematic image ana lysis, characterization and inte-
gration of numerous clinical and mammographic
variables [2], which is a difficult and error-prone
task for physicians. This leads to low positive
predictive value of imaging interpretation [3].
The integration of computer models into the
radiological imaging interpretation process can
increase the accuracy of image interpretation.
There are two broad categories of computer
models in breast cancer diagnosis: computer-
aided detection (CADe) and computer-aided
diagnostic (CADx) models. CADe models are
computerized tools that assist radiologists in
locating and identifying possible abnormalities
in radiologic images, leaving the interpretation of
the abnormality to the radiologist [4]. The poten-
tial for CADe models to improve detection of
cancer has been investigated in several retrospec-
tive studies [5–8] as well as carefully controlled
prospective studies [9–12]. For a review of CADe
studies, the reader is referred to recent review
articles by Hadjiiski et al. [13] and Nishikawa [14].
CADx models are decision aids for radiologists
characterizing findings from radiologic images
(e.g., size, contrast and shape) identified either
by a radiologist or a CADe model [15]. CADx
models have been demonstrated to increase the
accuracy of mammography interpretation in sev-
eral studies. Encouraged by promising results in
mammography interpretation, numerous CADx
models are being developed to help in breast US
and MRI interpretation.
There are two reviews of CADx models, but
neither are comprehensive in nature. The first,
by Elter and Horsch, focuses on CADx models
in mammography interpretation, but not in US
and MRI, and concentrates on technical aspects
of model development rather than more clini-
cally relevant considerations [16]. The second,
by Dorrius and van Ooijen, focuses on MRI
CADx models [17]. Here we provide a compre-
hensive review for mammography, US and MRI
CADx models in breast cancer diagnosis. We
start by summarizing CADx models proposed
for mammography interpretation. We then
describe CADx models in US and MRI. We
conclude by discussing several common limita-
tions of existing research on CADx models and
provide possible future research directions.
Mammography CADx models
Early work involving CADx models in mammo-
graphy interpretation dates back to 1993. A sum-
mary list for primary mammography CADx
models is presented in Table 1.
10.2217/IIM.10.24 © 2010 Future Medicine Ltd
Imaging Med. (2010) 2(3), 313–323
ISSN 1755-5191
313
For reprint orders, please contact: reprints@futuremedicine.com�
REVIEW Ayer, Ayvaci, Liu, Alagoz & Burnside
Table 1. Summary of computer-aided diagnostic models in
mammography interpretation.
Study (year)
Model
AUC
Ref.
Size of
dataset (n)
107
240
110
500
2100
253
115
419
62,219
62,219
62,219
151
[26]
Jiang et al. (1996)
[31]
Markopoulos et al. (2001)
[35]
Huo et al. (2002)
[37]
Floyd et al. (2000)
[38]
Elter et al. (2007)
[34]
Chan et al. (1999)
[41]
Gupta et al. (2006)
[42]
Wang et al. (1999)
[43]
Chhatwal et al. (2009)
[44]
Burnside et al. (2009)
[45]
Ayer et al. (2010)
[46]
Bilska-Wolak et al. (2005)
ANN: Artificial neural network; AUC: Area under the curve; BN: Bayesian network; CBR: Case-based
reasoning; DT: Decision tree; LDA: Linear discriminant ana lysis; LDC: Linear discriminant classifier;
LR: Logistic regression; LRbC: Likelihood ratio-based classifier.
0.92
0.937
0.96
0.83
0.87/0.89
0.91
0.92
0.886
0.963
0.960
0.965
0.88
ANN
ANN
ANN
CBR
DT/CBR
LDC
LDA
BN
LR
BN
ANN
LRbC
Reader
study
Yes
Yes
Yes
No
No
Yes
No
No
Yes
Yes
Yes
No
Early work of CADx research used artificial
neural networks (ANNs) and Bayesian networks
(BNs). The first CADx model was proposed by
Wu et al., who developed an ANN to classify
lesions detected by radiologists as malignant or
benign [18]. They demonstrated that their sim-
ple ANN, which was built using 14 radiologist-
extracted mammography features and trained on
a small set of data, achieved higher area under
the curve (AUC) of the receiver operating char-
acteristic (ROC) curve than a group of attending
radiologists without computer aid (0.89 vs 0.84).
Baker et al. later built more complex ANN mod-
els, where the inputs included Breast Imaging
Reporting and Data System (BI-RADS) descrip-
tors as well as variables related to the patient’s
medical history [19]. Their approach was later
extended and evaluated by others [20–23]. Fogel
et al. also built one of the early ANN models
that prospectively examined suspicious masses as
a second opinion to radiologists [24]. Kahn et al.
developed one of the first BN models to classify
mammographic lesions as benign and malignant
[25]. They used radiologist-extracted mammo-
graphy features as the input to their model and
demonstrated that BNs had a potential to help
radiologists making diagnostic decisions.
Jiang et al. trained an ANN to differentiate
malignant and benign clustered microcalcifica-
tions [26]. The microcalcifications were initially
identified by the radiologists and eight features
of these microcalcifications were automatically
extracted by an image-processing algorithm.
The training and testing data included 107 cases
(40 malignant) from 53 patients. This retrospec-
tive study only included microcalcifications that
underwent biopsy. Five radiologists participated
in the observer study. ROC ana lysis was used
to assess performance. The average cumula-
tive AUC values for the ANN and the radiolo-
gists were 0.92 and 0.89, respectively. While
the cumulative AUCs did not have a signifi-
cant difference (p = 0.22), the comparison of
AUCs over the 0.90 sensitivity threshold yielded
statistically significant differences (p < 0.05).
Jiang et al. later extended this model to classify
lesions as malignant or benign for multiple-view
mammograms [27]. They found that the use of
a CADx model decreased the number of biop-
sied benign lesions while increasing the biopsy
recommendations for malignant clusters. In a
follow-up study, Jiang et al. demonstrated that,
in addition to its diagnostic power, their ANN
model had the potential to reduce the variabil-
ity among radiologists in the interpretation of
mammograms [28]. In another study, they com-
pared their CADx model with independent dou-
ble readings on 104 mammograms (46 malig-
nant) containing clustered microcalcifications
and reported more significant improvements in
the ROC performance when the CADx model
was used as compared with the independent
double readings [29]. More recently, Rana et al.
applied the CADx model developed by Jiang
et al. on screen-film mammograms [26,27] to full-
field digital mammograms [30]. They concluded
that their CADx model maintained consistently
high performance in classifying calcifications in
full-field digital mammograms without requir-
ing substantial modifications from its initial
development on screen-film mammograms.
Markopoulos et al. compared three radio-
logists’ diagnostic accuracies with or with-
out computer aid [31]. The computer ana lysis
utilized an ANN in diagnosis of clustered
microcalcifications on mammograms. This
retrospective study included 240 suspicious
microcalcifications (108 malignant), which
were identified by radiologists and extracted
by an image-processing algorithm. The inputs
to the ANN included eight features of the cal-
cifications. Biopsy was the reference standard.
The AUC of the CADx was 0.937, which was
significantly higher than that of the physician
with the highest performance (AUC = 0.835,
p = 0.012). The authors concluded that CADx
models also have the potential to help improve
the diagnostic accuracy of radiologists.
Huo et al. also used ANNs to classify mass
lesions detected on screen-film mammograms
[32,33]. They automated the feature extraction proc-
ess to reduce the intra-observer variability [28,34].
314
Imaging Med. (2010) 2(3)
future science group
�
Computer-aided diagnostic models in breast cancer screening REVIEW
In a follow-up study, Huo et al. used different
sets of data for training and testing instead of
a single database [35]. Their database included
50 biopsy-proven malignant masses, 50 biopsy-
proven benign masses and ten cysts proved by
fine needle aspiration. The inputs to the ANN
included four characteristics of masses (mar-
gin, sharpness, density and texture) that were
automatically extracted by an image processing
algorithm. When the CADx model was used,
the average AUC of the radiologists increased
from 0.93 to 0.96 (p < 0.001), demonstrating
the generalizability of CADx models to distinct
datasets. More recently, Li et al. converted the
CADx model developed by Huo et al. on screen-
film mammograms to apply to full-field digital
mammograms [36]. They evaluated the per-
formance of this CADx model using the AUC
at various stages of the conversion process and
concluded that CADx models had a potential
to aid physicians in the clinical interpretation
of full-field digital mammograms.
Floyd et al. proposed a case-based reasoning
(CBR) approach, in which the classification is
based on the ratio of the matched malignant
cases to total matches in the database [37]. The
primary advantage of the CBR method over
an ANN is the transparent reasoning process
that leads to the system’s diagnosis. However, a
key limitation of CBR is that a new case might
not have any match in the database. This CBR
ana lysis included 500 (174 malignant) cases.
Of these 500 cases, 232 were masses alone, 192
were microcalcifications alone and 29 were com-
binations of masses and associated microcalci-
fications. The inputs to the CBR included ten
features from the BI-RADS lexicon (five mass
descriptors and five calcification descriptors) and
a descriptor from clinical data. Biopsy was the
reference standard. Two radiologists were asked
to describe each lesion using the BI-RADS lexi-
con. The input dataset contained both retrospec-
tive (206 cases) and prospective (194 cases) data.
The performance of the CBR model was com-
pared with that of an ANN. While the ANN
slightly outperformed the CBR (AUC = 0.86
vs 0.83, respectively), the study did not report
statistical significance of this difference.
Elter et al. evaluated two novel CADx
approaches that predicted breast biopsy outcomes
[38]. The study retrospectively analyzed cases that
contained masses or calcifications but not both.
The dataset included 2100 masses (1045 malig-
nant) and 1359 calcifications (610 malignant)
that were extracted from mammograms in a pub-
lic database and double reviewed by radiologists.
The positive cases included histologically proven
cancers, while negative cases were followed up
for a 2-year period. The inputs to the CADx
model included patient age and five features from
the BI-RADS lexicon (two mass descriptors and
three calcification descriptors). Elter et al. used
two types of CADx systems: a decision tree
and a CBR. An ANN was also implemented to
compare its performance to that of the two pro-
posed models. The models were evaluated based
on ROC ana lysis. Contrary to the findings by
Floyd et al. [37], they found that the CBR out-
performed the ANN (AUC = 0.89 vs 88, respec-
tively, p < 0.001), while the ANN performed bet-
ter than the decision tree (AUC = 0.88 vs 0.87,
respectively, p < 0.001). The authors concluded
that both systems could potentially reduce the
number of unnecessary biopsies with more
accurate prediction of breast biopsy outcomes.
However, the differences in AUC performances
were small, raising the possibility that they may
not be clinically significant.
Chan et al. retrospectively evaluated the
effects of a linear discriminant classifier on
radiologists’ characterization of masses [34]. The
dataset included 253 mammograms (127 malig-
nant). Biopsy was the reference standard. The
findings were initially identified by a radiologist
and 41 features of these findings (texture and
morphologic features) extracted by an image-
processing algorithm were used as inputs to the
linear discriminant classifier. Six reading radio-
logists evaluated the mammograms with and
without CADx. The classification performance
was evaluated by ROC ana lysis. The average
AUC of the reading radiologists without CADx
was 0.87 and improved to 0.91 with CADx
(p < 0.05). Hadjiiski et al. performed similar
studies to evaluate a CADx model and par-
ticularly investigated the extent of increase in
diagnostic accuracy when more mammographic
information was available [39,40]. Specifically,
they evaluated two scenarios: the increase in the
performance of CADx when trained on serial
mammograms [39] and the increase in the per-
formance of CADx when trained with interval
change ana lysis, which used interval change
information extracted from prior and current
mammograms [40]. For both scenarios, they
reported superior AUCs for the radiologists with
CADx when compared with the radiologists
without CADx (for the first scenario AUC = 0.85
vs 0.79, respectively, p = 0.005; and for the sec-
ond scenario AUC = 0.87 vs 0.83, respectively,
p < 0.05) and, thus, a significant improvement
of the radiologists’ diagnostic accuracy.
future science group
www.futuremedicine.com
315
�
REVIEW Ayer, Ayvaci, Liu, Alagoz & Burnside
Gupta et al. retrospectively studied 115
biopsy-proven masses or calcification lesions
(51 malignant) using a linear discriminant
ana lysis (LDA)-based CADx model [41]. The
images and case records were obtained from
a public database. This study compared the
performance of the LDA while using differ-
ent descriptors for one mammographic view
and two mammographic views. The attending
radiologists described each abnormality using
BI-RADS descriptors and categories. The inputs
to the CADx model included patient age and
two features from the BI-RADS lexicon (mass
shape and mass margin). While the CADx with
two mammographic views outperformed that
with one mammographic view (AUC = 0.920
vs 0.881, respectively), the difference was not
statistically significant (p = 0.056).
Wang et al. built and evaluated three BNs
[42]. One of the BNs was constructed based on a
total of 13 mammographic features and patients’
characteristics. The other two BNs were hybrid
classifiers, one of which was constructed by
averaging the outputs from two subnetworks
of mammographic-only or non-mammographic
features. The third classifier used logistic regres-
sion (LR) to compute the outputs from the same
subnetworks. This retrospective study included
419 cases (92 malignant). The verification of
positive cases included biopsy and/or surgical
reports, while negative cases were followed up
for at least a 2-year period. The input features
included four mammographic findings and nine
descriptors from clinical data. The features were
manually extracted by radiologists. The AUC
for the BN that incorporated all 13 features was
0.886 and the AUCs for the BNs that included
only mammographic features and patient char-
acteristics were 0.813 and 0.713, respectively.
The BN that included the full feature set was
significantly better than both of the hybrid BNs
(p < 0.05).
Recently, Chhatwal et al. [43] and Burnside
et al. [44] developed a LR and BN, respectively,
based on a consecutive dataset from a breast
imaging practice consisting of 62,219 mammog-
raphy records (510 malignant). The input fea-
tures included 36 variables based on BI-RADS
descriptors for masses, calcifications, breast
density, associated findings and patients’ clini-
cal descriptors. The input dataset was recorded
in the national mammography database format,
which allowed the use of these models in other
healthcare institutions. Contrary to most stud-
ies in the literature, they included the nonbiop-
sied mammograms in their training dataset and
used cancer registries as the reference standard
instead of the biopsy results. They analyzed the
performance of the CADx models using ROC
ana lysis and concluded that their CADx models
performed better than that of the radiologists
in aggregate (AUCs = 0.963 and 0.960 for LR
and BN, respectively, vs 0.939 for the radio logist;
p < 0.05). More recently, Ayer et al. developed
an ANN model using the same dataset and
demonstrated that the ANN model achieved
slightly a higher AUC (0.965) than that of the
LR and BN models as well as the radiologists
[45]. Additionally, Ayer et al. extended the per-
formance ana lysis of the CADx models from
discrimination (classification) to calibration
metrics, which assessed the ability of this ANN
model to accurately predict the cancer risk for
individual patients.
Bilska-Wolak et al. conducted a preclinical
evaluation of a previously developed CADx
model, a likelihood ratio-based classifier, on
a new set of data [46]. The model retrospec-
tively evaluated 151 new and independent
cases (42 malignant). Biopsy was the reference
standard. Suspicious masses were detected and
described by an attending radiologist using
16 different features from the BI-RADS lexi-
con and patient history. The authors evaluated
the CADx model based on ROC ana lysis and
sensitivity statistics. The average AUC was 0.88.
The model achieved 100% sensitivity at 26%
specificity. The results were compared with an
ANN model created using the same datasets.
The AUC of the ANN was lower than that of the
likelihood ratio-based classifier. Bilska-Wolak
et al. concluded that their CADx model showed
promising results that could reduce the number
of false-positive mammograms.
US CADx models
Ultrasound imaging is an adjunct to diagnostic
mammography, where CADx models could be
used for improving diagnostic accuracy. CADx
models developed for US scans date back to late
1990s. In this section, we review studies that
apply CADx systems to breast sonography or
US-mammography combination in distinguish-
ing malignant from benign lesions. A sum-
mary list for the primary US CADx models is
presented in Table 2.
Giger et al. classified malignant lesions in a
database of 184 digitized US images [47]. Biopsy,
cyst aspiration or image interpretation alone were
used to confirm benign lesions, whereas malig-
nancy was proven at biopsy. The authors utilized
an LDA model to differentiate between benign
316
Imaging Med. (2010) 2(3)
future science group
�
Computer-aided diagnostic models in breast cancer screening REVIEW
and malignant lesions using five computer-
extracted features based on lesion shape and mar-
gin, texture, and posterior acoustic attenuation
(two features). ROC ana lysis yielded AUCs of
0.94 for the entire database and 0.87 for the data-
base that only included biopsy- and cyst-proven
cases. The authors concluded that their ana lysis
demonstrated that computerized ana lysis could
improve the specificity of breast sonography.
Chen et al. developed an ANN to classify
malignancies on US images [48]. A physician
manually selected sub-images corresponding to
a suspicious tumor region followed by compu-
terized ana lysis of intensity variation and tex-
ture information. Texture correlation between
neighboring pixels was used as the input to the
ANN. The training and testing dataset included
140 biopsy-proven breast tumors (52 malig-
nant). The performance was assessed by AUC,
sensitivity and specificity metrics, which yielded
an AUC of 0.956 with 98% sensitivity and 93%
specificity at a threshold level of 0.2. The authors
concluded that their CADx model was useful in
distinguishing benign and malignant cases, yet
also noted that larger datasets could be used to
improve the performance.
Later, Chen et al. improved on a previous
study [48] and devised an ANN model com-
posed of three components: feature extraction,
feature selection, and classification of benign and
malignant lesions [49]. The study used two sets of
biopsy-proven lesions; the first set with 160 dig-
itally stored lesions (69 malignant) and the sec-
ond set with 111 lesions (71 malignant) in hard-
copy images that were obtained with the same
US system. Hard-copy images were digitized
using film scanners. Seven morphologic features
were extracted from each lesion using an image-
processing algorithm. Given the classifier, for-
ward stepwise regression was employed to define
the best performing features. These features were
used as inputs to a two-layer feed-forward ANN.
For the first set, the ANN achieved an AUC of
0.952, 90.6% sensitivity and 86.6% specificity.
For the second set, the ANN achieved an AUC
of 0.982, 96.7% sensitivity and 97.2% specifi-
city. The ANN model trained on each dataset
was demonstrated to be statistically extendible
to other datasets at a 5% significance level. The
authors concluded that their ANN model was an
effective and robust approach for lesion classifi-
cation, performing better than the counterparts
published earlier [47,48].
Horsch et al. explored three aspects of an LDA
classifier that was based on automatic segmenta-
tion of lesions and automatic extraction of lesion
Table 2. Summary of computer-aided diagnostic models in
ultrasound interpretation.
Study (year)
Model
AUC
Size of
dataset (n)
184
140
160/111
400
102
1046
717
67
Giger et al. (1999)
Chen et al. (1999)
Chen et al. (2003)
Horsch et al (2002)
Sahiner et al. (2004)
Drukker et al. (2008)
Horsch et al. (2006)
Sahiner et al. (2009)
ANN: Artificial neural network; AUC: Area under the curve; BNN: Bayesian neural network;
LDA: Linear discriminant analysis.
0.94
0.956
0.952/0.982
0.87
0.92
0.90
0.91
0.95
LDA
ANN
ANN
LDA
LDA
BNN
BNN
LDA
Reader
study
No
No
No
No
Yes
Yes
Yes
Yes
Ref.
[47]
[48]
[49]
[50]
[51]
[52]
[53]
[54]
shape, margin, texture and posterior acoustic
behavior [50]. The study was conducted using a
database of 400 cases with 94 malignancies, 124
complex cysts and 182 benign lesions. The refer-
ence standard was either biopsy or aspiration. First,
marginal benefit of adding a feature to the LDA
model was investigated. Second, the performance
of the LDA model in distinguishing carcinomas
from different benign lesions was explored. The
AUC values for the LDA model were 0.93 for dis-
tinguishing carcinomas from complex cysts and
0.72 for differentiating fibrocystic disease from
carcinoma. Finally, eleven independent trials of
training and testing were conducted to validate
the LDA model. Validation resulted in a mean
AUC of 0.87 when computer-extracted features
from automatically delineated lesion margins were
used. There was no statistically significant dif-
ference between the best two- and four-feature
classifiers; therefore, adding features to the LDA
model did not improve the performance.
Sahiner et al. investigated computer vision
techniques to characterize breast tumors on
3D US volumetric images [51]. The dataset was
composed of masses from 102 women who
underwent either biopsy or fine-needle aspira-
tion (56 had malignant masses). Automated
mass segmentation in 2D and 3D, as well as
feature extraction followed by LDA, were imple-
mented to obtain malignancy scores. Stepwise
feature selection was employed to reduce eight
morphologic and 72 texture features into a best-
feature subset. An AUC of 0.87 was achieved
for the 2D-based classifier, while the AUC for
the 3D-based classifier was 0.92. There was no
statistically significant difference between the
two classifiers (p = 0.07). The AUC values of the
four radiologists fell in the range of 0.84 to 0.92.
Comparing the performance of their model to
that of radiologists, the difference was not sta-
tistically significant (p = 0.05). However, the
future science group
www.futuremedicine.com
317
�
REVIEW Ayer, Ayvaci, Liu, Alagoz & Burnside
partial AUC for their model was significantly
higher than those of the three radiologists
(p < 0.03, 0.02 and 0.001).
Drukker et al. used various feature segmen-
tation and extraction schemes as inputs to a
Bayesian neural network (BNN) classifier with
five hidden layers [52]. The purpose of the study
was to evaluate a CADx workstation in a realistic
setting representative of clinical diagnostic breast
US practice. Benign or malignant lesions that
were verified at biopsy or aspiration, as well as
those determined through imaging characteristics
on US scans, MR images and mammograms, were
used for the ana lysis. The authors included non-
biopsied lesions in the dataset to make the series
consecutive, which more accurately reflects clini-
cal practice. The inputs to the network included
lesion descriptors consisting of the depth:width
ratio, radial gradient index, posterior acoustic sig-
nature and autocorrelation texture feature. The
output of the network represented the probabil-
ity of malignancy. The study was conducted on a
patient population of 508 (101 had breast cancer)
with 1046 distinct abnormalities (157 cancer-
ous lesions). Comparing the current radiology
practice with the CADx workstation, the CADx
scheme achieved an AUC of 0.90, corresponding
to 100% sensitivity at 30% specificity, while radi-
ologists performed with 77% specificity for 100%
sensitivity when only nonbiopsied lesions were
included. When only biopsy-proven lesions were
analyzed, computerized lesion characterization
outperformed the radiologists.
In routine clinical practice, radiologists often
combine the results from mammography and US,
if available, when making diagnostic decisions.
Several studies demonstrated that CADx could
be useful in the differentiation of benign findings
from malignant breast masses when sonographic
data are combined with corresponding mammo-
graphic data. Horsch et al. evaluated and com-
pared the performance of five radiologists with
different expertise levels and five imaging fellows
with or without the help of a BNN [53]. The BNN
model utilized a computerized segmentation of
the lesion. Mammographic features used as the
input included spiculation, lesion shape, margin
sharpness, texture and gray level. Sonographic
input features included lesion shape, margin,
texture and posterior acoustic behavior. All fea-
tures were automatically extracted by an image-
processing algorithm. This retrospective study
examined a total of 359 (199 malignant) mam-
mographic and 358 (67 malignant) sonographic
images. Additionally, 97 (39 malignant) multimo-
dality cases (both mammogram and sonogram)
were used for testing purposes only. Biopsy was
the reference standard. The performances of each
radiologist/imaging fellow or pair of observers
were quantified by the AUC, sensitivity and spe-
cificity metrics. Average AUC without BNN was
0.87 and with BNN was 0.92 (p < 0.001). The
sensitivities without and with BNN were 0.88 and
0.93, respectively (p = 0.005). There was not a
significant difference in specificities without and
with BNN (0.66 vs 0.69, p = 0.20). The authors
concluded that the performance of the radiologists
and imaging fellows increased significantly with
the help of the BNN model.
In another multimodality study, Sahiner et al.
investigated the effect of a multimodal CADx
system (using mammography and US data) in
discriminating between benign and malignant
lesions [54]. The dataset for the study consisted of
13 mammography features (nine morphologic,
three spiculation and one texture) and eight 3D
US features (two morphologic and six texture)
that were extracted from 67 biopsy-proven masses
(35 malignant). Ten experienced readers first gave
a malignancy score based on mammography only,
then re-evaluated based on mammography and
US combined, and were finally allowed to change
their minds given the CADx system’s evaluation
of the mass. The CADx system automatically
extracted the features, which were then fed into
a multimodality classifier (using LDA) to give a
risk score. The results were compared using ROC
curves, which suggested statistically significant
improvement (p = 0.05) when the CADx system
was consulted (average AUC = 0.95) over read-
ers’ assessment of combined mammography and
US without the CADx (average AUC = 0.93).
Sahiner et al. concluded that a CADx system
combining the features from mammography and
US may have the potential to improve radiologist’s
diagnostic decisions [54].
As discussed previously, a variety of sono-
graphic features (texture, margin and shape) are
used to classify benign and malignant lesions.
2D/3D Doppler imaging provides additional
advantages in classification when compared
with grayscale, by demonstrating breast lesion
vascularity. Chang et al. extracted features of
tumor vascularity from 3D power Doppler US
images of 221 lesions (110 benign) and devised
an ANN to classify lesions [55]. The study dem-
onstrated that CADx, using 3D power Doppler
imaging, can aid in the classification of benign
and malignant lesions.
In addition to the aforementioned studies,
there are other works that developed and evalu-
ated CADx systems in differentiating between
318
Imaging Med. (2010) 2(3)
future science group
�
Computer-aided diagnostic models in breast cancer screening REVIEW
benign and malignant lesions. Joo et al. devel-
oped an ANN that was demonstrated to have
potential to increase the specificity of US char-
acterization of breast lesions [56]. Song et al.
compared an LR and an ANN in the context of
differentiating between malignant and benign
masses on breast sonograms from a small dataset
[57]. There was no statistically significant differ-
ence between the performances of the two meth-
ods. Shen et al. investigated the statistical cor-
relation between the computerized sonographic
features, as defined by BI-RADS, and the signs
of malignancy [58]. Chen and Hsiao evaluated
US-based CADx systems by reviewing the meth-
ods used in classification [59]. They suggested the
inclusion of pathologically specific tissue-and
hormone-related features in future CADx sys-
tems. Gruszauskas et al. examined the effect of
image selection on the performance of a breast
US CADx system and concluded that their auto-
mated breast sonography classification scheme
was reliable even with variation in user input [60].
Recently, Cui et al. published a study focusing on
the development of an automated method seg-
menting and characterizing the breast masses on
US images [61]. Their CADx system performed
similarly whether it used automated segmenta-
tion or an experienced radiologist’s segmentation.
In a recent study, Yap et al. designed a survey to
evaluate the benefits of computerized processing
of US images in improving the readers’ perform-
ance of breast cancer detection and classification
[62]. The study demonstrated marginal improve-
ments in classification when computer-processed
US images alongside the originals are used in
distinguishing benign from malignant lesions.
MRI CADx models
Dynamic contrast-enhanced MRI of the breast
has been increasingly used in breast cancer
evaluation and has been demonstrated to have
potential to improve breast cancer diagnosis. The
major advantage of MRI over other modalities is
its ability to depict both morphologic and physio-
logic (kinetic enhancement) information [63].
Despite the advantages of MRI, it is a technology
that is continuously evolving and is not currently
cost effective for screening the general population
[64,65]. Nevertheless, breast MRI is promising in
terms of its high sensitivity, especially for high-
risk young women with dense breasts. However,
specificity has been highly variable in detection
of breast cancer [17]. As a way of improving spe-
cificity, CADx models to aid discrimination of
benign from malignant lesions in MRI imaging
would be valuable. There are numerous CADx
studies based on breast MRI. Generally, both
morphologic and kinetic (enhancement) features
are used in these studies to predict benign versus
malignant breast lesions. In this section of the
article, we only discuss the recent articles (pub-
lished after 2003) that exemplify distinct aspects
of breast MRI CADx research. A summary list
for the primary MRI CADx models is presented
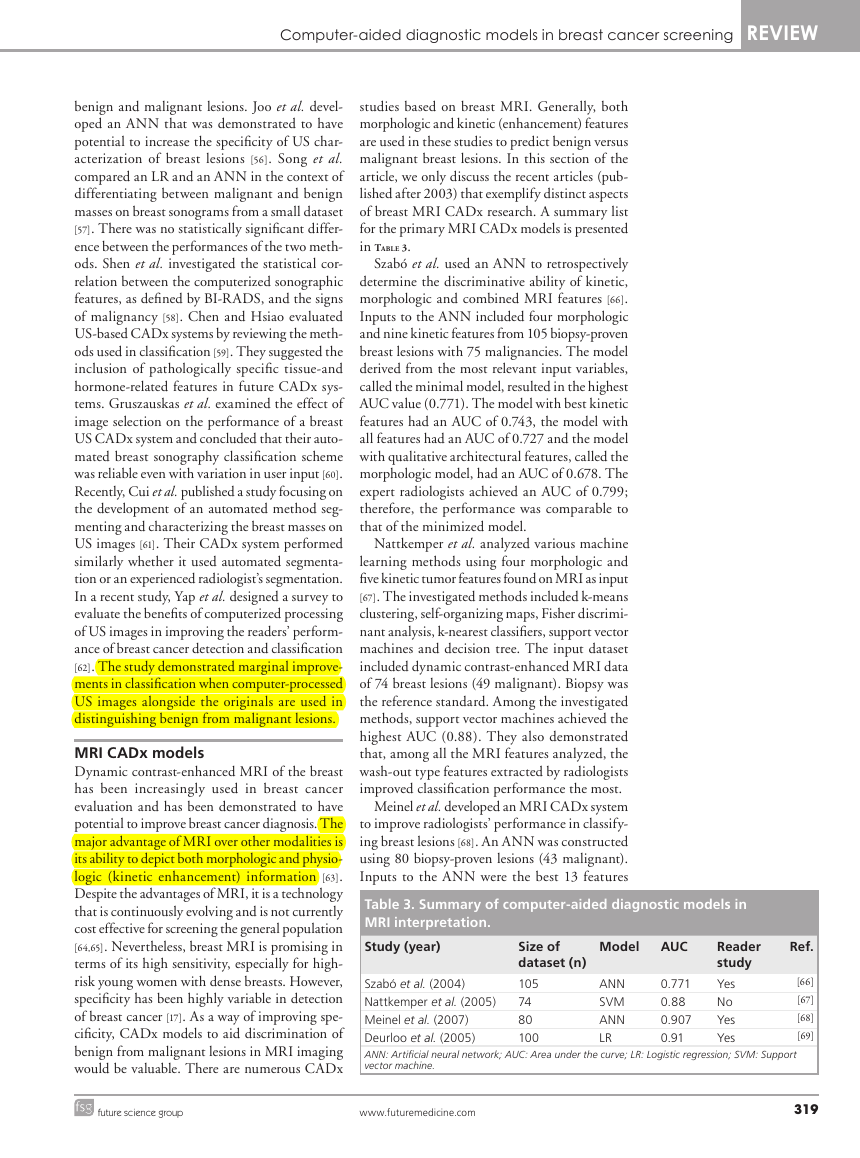
in Table 3.
Szabó et al. used an ANN to retrospectively
determine the discriminative ability of kinetic,
morphologic and combined MRI features [66].
Inputs to the ANN included four morphologic
and nine kinetic features from 105 biopsy-proven
breast lesions with 75 malignancies. The model
derived from the most relevant input variables,
called the minimal model, resulted in the highest
AUC value (0.771). The model with best kinetic
features had an AUC of 0.743, the model with
all features had an AUC of 0.727 and the model
with qualitative architectural features, called the
morphologic model, had an AUC of 0.678. The
expert radiologists achieved an AUC of 0.799;
therefore, the performance was comparable to
that of the minimized model.
Nattkemper et al. analyzed various machine
learning methods using four morphologic and
five kinetic tumor features found on MRI as input
[67]. The investigated methods included k-means
clustering, self-organizing maps, Fisher discrimi-
nant ana lysis, k-nearest classifiers, support vector
machines and decision tree. The input dataset
included dynamic contrast-enhanced MRI data
of 74 breast lesions (49 malignant). Biopsy was
the reference standard. Among the investigated
methods, support vector machines achieved the
highest AUC (0.88). They also demonstrated
that, among all the MRI features analyzed, the
wash-out type features extracted by radiologists
improved classification performance the most.
Meinel et al. developed an MRI CADx system
to improve radiologists’ performance in classify-
ing breast lesions [68]. An ANN was constructed
using 80 biopsy-proven lesions (43 malignant).
Inputs to the ANN were the best 13 features
Table 3. Summary of computer-aided diagnostic models in
MRI interpretation.
Study (year)
Model
AUC
Size of
dataset (n)
105
74
80
100
Szabó et al. (2004)
Nattkemper et al. (2005)
Meinel et al. (2007)
Deurloo et al. (2005)
ANN: Artificial neural network; AUC: Area under the curve; LR: Logistic regression; SVM: Support
vector machine.
0.771
0.88
0.907
0.91
ANN
SVM
ANN
LR
Reader
study
Yes
No
Yes
Yes
Ref.
[66]
[67]
[68]
[69]
future science group
www.futuremedicine.com
319
�
REVIEW Ayer, Ayvaci, Liu, Alagoz & Burnside
from a set of 42, based on lesion shape, texture
and enhancement kinetics information. The per-
formance was assessed by comparison of AUC
values from five human readers diagnosing the
tumor with and without the help of the CADx
system. When only the first abnormality shown
to human readers was included, ROC ana lysis
yielded AUCs of 0.907 with ANN assistance
and 0.816 without the assistance. The difference
was statistically significant (p < 0.011); therefore,
Meinel et al. demonstrated that their ANN model
improves the performance of human readers.
Deurloo et al. combined the clinical assessment
of clinically and mammographically occult breast
lesions by radiologists with computer-calculated
probability of malignancy of each lesion into an
LR model [69]. Inputs to the LR model included
the four best features from a set of six morphologic
and three temporal features. Either biopsy-proven
lesions or lesions showing transient enhancement
were included in the study. The difference between
the performance of clinical readings (AUC = 0.86)
and computerized ana lysis (AUC = 0.85) was not
statistically significant (p = 0.99). However, the
combined model performed significantly higher
(AUC = 0.91, p = 0.03) when compared with
clinical reading without computerized ana lysis.
The results demonstrated how computerized ana-
lysis could complement clinical interpretation of
magnetic resonance images.
There are several other studies that addressed
the use of CADx systems in MRI of the
breast. Williams et al. evaluated the sensitiv-
ity of computer-generated kinetic features from
CADstream, the first CADx system for breast
MRI, for 154 biopsy-proven lesions (41 malig-
nant) [70]. The study suggested that computer-
aided classification improved radiologists’ per-
formance. Lehman et al. compared the accuracy
of breast MRI assessments with and without the
same software, CADstream [71]. They concluded
that the software may improve the accuracy of
radiologists’ interpretation; however, the study
was conducted on a small set of 33 lesions (nine
malignant). Nie et al. investigated the feasibil-
ity of quantitative ana lysis of MRI images [72].
Morphology/texture features of breast lesions
were selected by an ANN and used in the classi-
fication of benign and malignant lesions. Baltzer
et al. investigated the incremental diagnostic
value of complete enhancing lesions using a
CADx model [73]. The study reported improve-
ment in specificity with no statistical significance.
In a different study, Baltzer et al. investigated
both automated and manual measurement meth-
ods to assess contrast enhancement kinetics [74].
They analyzed and compared evaluation of con-
trast enhancements via curve-type assessment by
radiologists, region of interest and CADx. The
methods proved diagnostically useful although
no statistically significant difference was found.
Future perspective
There have been significant advances in CADx
models in the last 20 years. However, several
issues remain open for future researchers. First
and most notably, almost all of the existing
CADx models are trained and tested on retro-
spectively collected cases that may not represent
the real clinical practice. Large prospective stud-
ies are required to evaluate the performance of
CADx models in real life before employing them
in a clinical setting.
Second, an objective comparative perform-
ance evaluation of the existing CADx models
is difficult because the reported performances
depend on the dataset used in model build-
ing. One approach to a systematic performance
comparison would be to use large and consistent,
publicly available datasets for testing purposes.
However, although this approach will give some
idea about the realistic/comparable performances
of the CADx systems, it would not be completely
accurate because a CADx model performing the
best on one dataset might be outperformed by
another CADx model on another dataset.
Third, a frequently ignored issue in CADx
model development is the clinical interpretability
of the model. Aspects of the CADx model that
allow clinical interpretations significantly influ-
ence the acceptance of the CADx model by the
physicians. Most of the existing CADx models
are based on ANNs. Although ANNs are pow-
erful in terms of their predictive abilities, their
parameters do not carry any real-life interpreta-
tion, hence, they are often referred to as ‘black
boxes’. Other models such as LR, BN or CBR
allow direct clinical interpretation. However, the
number of such studies is significantly limited as
compared with ANN models.
Fourth, performance assessment of the CADx
models are usually limited to discrimination (clas-
sification) metrics (e.g., sensitivity, specificity and
AUC). On the other hand, the accuracy of risk
prediction for individual patients, referred to as
calibration, is often ignored. Although discrimi-
nation assesses the ability to correctly distinguish
between benign and malignant abnormalities, it
does not tell much about the accuracy of risk pre-
diction for individual patients [75]. However, clini-
cal decision-making usually involves decisions for
individual patients under uncertainty; therefore,
320
Imaging Med. (2010) 2(3)
future science group
�















 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc