Introduction
This article concerns a project about diffusion limited aggregation (or DLA). A DLA is a growing
object consisting of particles that diffuse and aggregate to the object, much like how flocks of snow are
created, and is simulated by a computer program. In such a simulation it is possible to determine some
of the properties that DLA's have. In this report I'll tell about how I've implemented the simulation and I
will try to discover some of the properties of DLA. Since I've made a somewhat non-trivial
implementation, I will spent a great amount of this article on it.
The DLA-algorithm
A DLA consists of particles that attach to each other. How they attach is described here.
First of all, we start with a complete empty space and put one particle somewhere in it. Now, a second
particle is 'fired' at an infinitely distance away from this particle. This new particle keeps moving
randomly in the empty space until it collides with the already placed particle. At this moment, the
particles attach to each other and are settled. Now a third particle is fired likewise, and the same
procedure repeats itself over and over.
Implementation
Simplifications
To simulate the continuous algorithm on a discrete device like a computer with a finite amount of time,
we need to make some changes to the algorithm. First of all; new particles aren't fired infinitely far
away from the object, but rather are fired just outside the bounding circle of the object. The place on
this circle is randomly chosen with a uniform distribution over the entire circle. Since the particle
would have reached this circle anyhow, the outcome of the algorithm is not changed by this limitation
(the time taken by the algorithm however is changed). Also, since we can't simulate a continuously
moving particle, we simulate it by making discrete steps with a certain distance.
Moreover, to make things speed up a little bit more, when particles aren't interested in our neat DLA
and start to drift away, we kill the particle at some distance and fire a new particle. This procedure
actually changes the chances for attaching to the particles very, very slightly. This is because the chance
for a new particle to get killed while reaching a certain particle in the DLA is for each particle in the
DLA different. Therefore, there are certain positions which become a little bit harder to reach. Since we
will see that particles aren't killed that often if we make the kill-distance large, we can do it
nevertheless.
Another simplification we do to speed things up is to let particles 'jump' when they get outside the
bounding circle of the DLA. Since, if the particle does not collide, the chances for the particle to reach
some point on a circle around itself are uniformly spread (this is actually not true for small circles on a
square lattice, as we will do later), therefore we can as well place it on a random position on the circle
immediately.
�
I wrote two programs which implemented the algorithm with the above simplifications. In the first one,
another simplification was made; particles were only able to move on a square lattice. Therefore,
besides that a particle cannot make a step in any direction, it can only make a step in horizontal or
vertical direction, and optionally it might move diagonally. This simplification makes implementing the
algorithm much easier, and will result in a better performance. In the second program, which is
discussed after the square lattice program, particles are off-lattice and can move in any direction. We
will see that the square-lattice simplification changes the shape of the lattice.
Implementation of the square lattice DLA
Both implementations were written in the C++ computer language. Like in the previous project (“The
Chirikov map”) I used the library SDL to do the graphics for me. I will only mention the most
interesting code or algorithms here, the program altogether can be find in the appendices.
The data structure is fairly simple in the square lattice case; it suffices to have a two-dimensional array
of integers of size n x n, where n is the maximum size that we want to reach. If we do not know the size
on forehand, we can dynamically grow the array as needed. The integers represents each position on the
square lattice. Each integer is initialized to zero, except for the element in the array which represent the
point in the middle. This point is initialized to the number 'minValue'. The value of each integer is
incremented when a particles 'ends' at the position that integer represents. When the integer has reached
'minValue', the particle on this position will actually become part of the DLA and new particles can
attach to this particle.
In a normal DLA, the value minValue will be one, which will cause particles to attach to the DLA the
first time they collide. However, if we give minValue some higher number, the randomness of the
algorithm will be 'averaged', and we can see the properties that the algorithm brings without having to
generate huge DLA's.
The function in which a new point is proposed and which contains the DLA algorithm is
DLALattice::CalculatePoint(..), which can be found just below. It is made in such a way that the
steps can be made with high performance; floating point calculations are avoided and used as few times
as possible and square roots are avoided by taking the square of the radii. The last interesting thing this
method implements is the use of a 'sticky value'. This value determines how much chance there is for a
particle to stick to the DLA. This parameter, which normally will have a value of 1 (=particles will
always stick, the value is actually pre-multiplied by RAND_MAX to avoid a floating point variable), can be
used to change the shape of the DLA as well.
�
void DLALattice::CalculatePoint(int &x, int &y) const {
GetRandomPointOnCircle(x, y, centerX, centerY, 1.1*(double) (radiusMax+5));
int radiusKillSquared = radiusKill * radiusKill;
int radiusJumpSquared = radiusJump * radiusJump;
bool stick=false;
do {
while(!HasNeighbour(x,y)) {
int choice = random() / (RAND_MAX / 4);
switch(choice) {
case 0: x++; break;
case 1: y++; break;
case 2: x--; break;
case 3: y--; break;
}
int r = (x-centerX)*(x-centerX) + (y-centerY)*(y-centerY);
while(r>radiusJumpSquared) {
if(r>radiusKillSquared) {
// Kill
GetRandomPointOnCircle(x, y, centerX, centerY, 1.1*(double)
(radiusMax+5));
} else {
// Jump
GetRandomPointOnCircle(x, y, x, y, sqrt(r)-radiusMax);
}
r = (x-centerX)*(x-centerX) + (y-centerY)*(y-centerY);
}
}
stick = (!useSticky) || random()
Implementation of the off-lattice DLA
The data structure that holds an off-lattice DLA is less trivial, compared to the square lattice case. In
this case, we do not assume particles are square-shaped, but will represent each of the particles as a
circle. Also, we will lose the simplification that particles could move only horizontally or vertically.
Particles are moving freely through our area now, making steps in any direction.
A simple method for implementing the circle collection
We cannot simple store each particle in one element of the n x n matrix as we did in the square lattice
case. However, with a minor change to the matrix, we can. Since
we know that each particle, represented as a circle, is of constant
diameter, say d, we can divide our area in squares of d x d and we
know that each square can hold one circle as a maximum, since the
circles are at least 2d away from each other. If we store each square
in an element in the n x n matrix, we're settled.
To see if two circles intersect, which is done continuously when we
fire a particle, we can check the square in which the new particle
would be placed. If there is a circle in this square, we're sure that
they will collide. If there isn't we check whether there are circles in
one of the nine neighboring squares. If so, we check for each circle
whether the distance between the centers of the new and existing
particles is less then 2d. If so, we know that they collide. If the
circle intersects with neither a circle in its square, nor intersects with a circle in the nine neighbors, we
can be sure that the circle does not intersect with any of the circles in our collection of circles.
Mapping of round particles to a grid
If we store the particles like described, we have approximately the same advantages and disadvantages
as in the square lattice case. The advantages are:
● O(1) worst-case time complexity for testing whether a specified circle intersects with one in the
collection (ie., it can be done in constant time)
● O(1) worst-case time complexity for inserting a new circle
● Fairly simple implementation
● It takes at most 9 intersection tests to see whether a specific circle intersects with one in the
collection
Limitations of this data structure include:
● Diameter of the circles has to be constant
● We cannot store intersecting circles in our data structure
● We have to declare the size of the grid on forehand, or change the size of the grid when it can not
hold its circles any longer
● Iterating all circles takes O(n2) time, where n is the width of the grid. Since not all squares will
contain a circle, this will usually mean we spent more time than necessary. In the case of our DLA
for example, if we already know that our DLA has a dimension of 1.5, the DLA will be of diameter
m(2/3) (m is mass of the DLA). Since our grid has to be at least of this size and iterating will cost
quadratic more, the iteration will cost O(m(4/3))
● The same calculation holds for the worst-case space requirement: this will also be of O(m(4/3)) in the
● To achieve the data structure's space requirement, we assume that we know that the circles will be
case of our DLA.
stored within a certain bounding box.
�
Since each newly fired particle is being tested for intersection with all other particles until it actually
intersects and is attached to the particle, the efficiency of testing the intersects seems to set the
performance of our program. Is this true?
The distance that each particle travels before hitting another particle grows, because of the growing
diameter of the bounding circle on which new particles are fired. Because of our jump and killing
strategy, particles tent to move towards the DLA. Later we will see that reaching the DLA costs
approximately O(m) steps1. Therefore, the insertion operation can cost O(m) time without changing the
time complexity of the entire algorithm.
Also, if we do not perform a circle-iterate operation more than once every m(1/3) insertions, the iteration
will not influence the algorithm. A circle-iteration operation occurs when we save the circles to a file or
when we enlarge the grid. Therefore, neither should we do more than once every m(1/3) particles. This is
not a problem though, since if we make the grid two times larger in horizontal and vertical direction
every time the DLA is about to overgrow the grid, we perform the operation once per 0,63m on
average, because the diameter of the bounding circle of the DLA will only be doubled every 0,63m
particles (and once every 0,63m insertions is less often than once every m(1/3) insertions, for all m>2
approximately). Therefore, the test for intersection does indeed set the performance for our program.
It is interesting to note that, if we would have increased the size of the grid every c particles, where c is
a constant, we would affect our time-complexity, since performing an O(m(4/3)) operation every c
insertions implies that inserting will cost O(m(4/3)), by which the insertion will cost more than the
number of steps we have to take each insertion.
Note that the above calculations are true in the square lattice case as well.
We can conclude from this that the above data structure is almost ideal for our problem. The only thing
that is not so ideal is the fact that we need O(m(4/3)) space. Considering the fact that simulating DLA's
containing one billion particles, which will probably be far beyond the maximum amount of particles
we can simulate considering the time this is going to cost, and considering the fact that a DLA of one
billion particles will cost (10^9)(4/3) = 1012 ≈ 1 terrabyte of space – and this is still more or less
technically possible, this is not our first problem.
In short: this data structure is ideal for our problem. I didn't use it however... Why not?? Well, I
preferred something... ehm.. more difficult. I tried to implement the circle collection in such a way that
some of the previously called disadvantages disappeared, without influencing the time complexity...
much. The data structure and intersection algorithm I used will actually perform better in some
situations In the upcoming paragraph I will describe this algorithm.
1 Since the direction of the steps are based on random numbers, this is not a worst case performance, but rather an average
performance. In worst case scenario, the algorithm will never end.
�
A less simple method for implementing the circle collection
In our second method we will use a completely different technique. In this implementation, the circles
are sorted and stored in an AVL-tree. To do so, we need an ordering in which circles that are closely
together remain closely together. This is 'more or less' achieved by ordering the circles
lexicographically first on ring and then on angle.
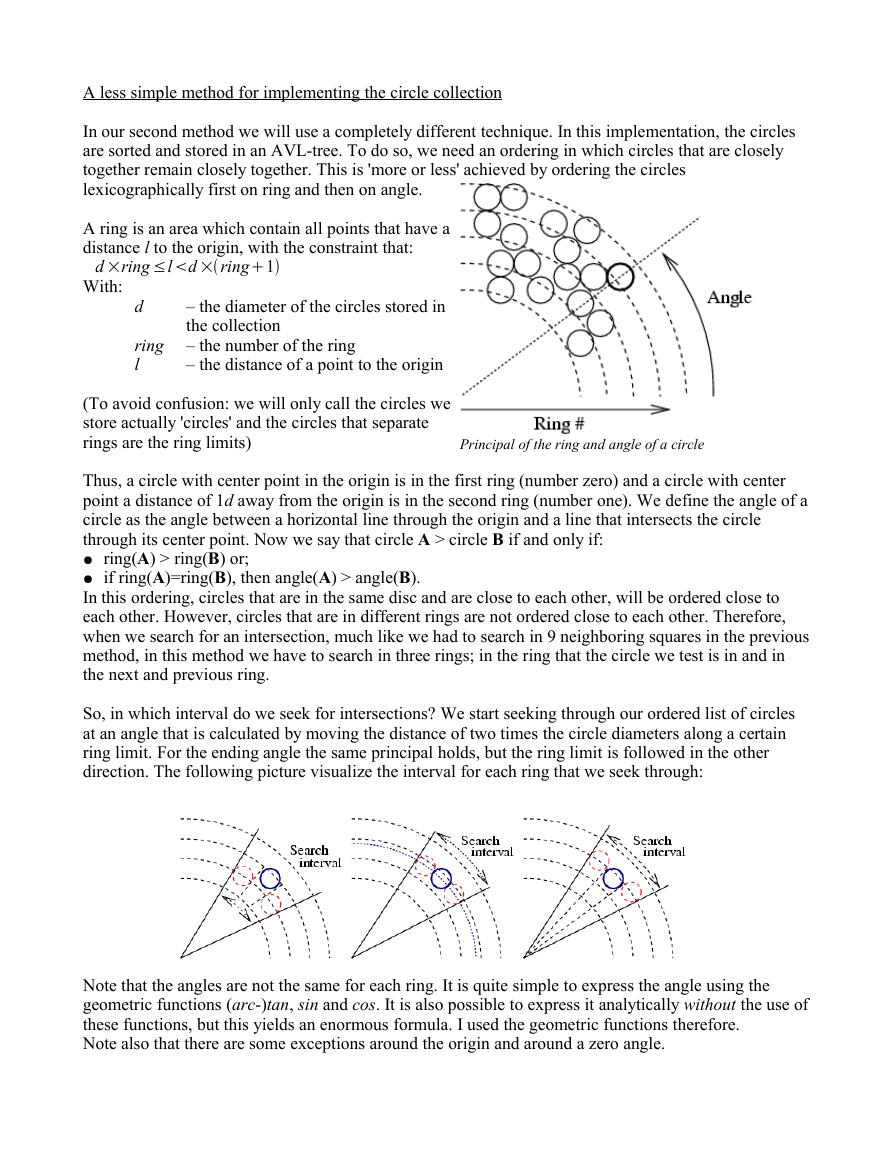
A ring is an area which contain all points that have a
distance l to the origin, with the constraint that:
d ×ring ≤ld ×ring1
With:
d
ring
l
– the diameter of the circles stored in
the collection
– the number of the ring
– the distance of a point to the origin
(To avoid confusion: we will only call the circles we
store actually 'circles' and the circles that separate
rings are the ring limits)
Principal of the ring and angle of a circle
Thus, a circle with center point in the origin is in the first ring (number zero) and a circle with center
point a distance of 1d away from the origin is in the second ring (number one). We define the angle of a
circle as the angle between a horizontal line through the origin and a line that intersects the circle
through its center point. Now we say that circle A > circle B if and only if:
● ring(A) > ring(B) or;
● if ring(A)=ring(B), then angle(A) > angle(B).
In this ordering, circles that are in the same disc and are close to each other, will be ordered close to
each other. However, circles that are in different rings are not ordered close to each other. Therefore,
when we search for an intersection, much like we had to search in 9 neighboring squares in the previous
method, in this method we have to search in three rings; in the ring that the circle we test is in and in
the next and previous ring.
So, in which interval do we seek for intersections? We start seeking through our ordered list of circles
at an angle that is calculated by moving the distance of two times the circle diameters along a certain
ring limit. For the ending angle the same principal holds, but the ring limit is followed in the other
direction. The following picture visualize the interval for each ring that we seek through:
Note that the angles are not the same for each ring. It is quite simple to express the angle using the
geometric functions (arc-)tan, sin and cos. It is also possible to express it analytically without the use of
these functions, but this yields an enormous formula. I used the geometric functions therefore.
Note also that there are some exceptions around the origin and around a zero angle.
�
We now have enough information to store the circles in an ALV-tree. Implementing an ALV-tree was
less trivial then I though (especially since I've implemented the tree avoiding any recursion in the
insertion and seek routines – for which I have no good reason though. It might be a little bit more
efficient). The resulting algorithm has the following properties:
● O(log n) worst-case time complexity for testing whether two particles intersect (this is caused by the
fact that we have to traverse the tree with height log n from root to bottom to find the first circle that
is higher than the start angle)
● O(log n) worst-case time complexity for inserting a new particle
● It takes at most 9 intersection tests to see whether a specified circle intersects with one in the
collection
● Space requirement is O(n)
● Iterating all circles takes O(n)
● In contrast to the first algorithm, we do not have to assume that the circles will be stored within a
certain bounding circle: we can insert a particle in the collection at a long distance from the origin
without affecting properties of the collection.
● In contrast to the first algorithm, we can store intersecting circles in our data structure. However, if
we do so, the 9 intersection tests-maximum is not hold any longer.
● Remark that using this method we can also make an intersection graph of n equal-sized circles in
O(n log n + i) (where i is the number of intersections) expected time.
So we gained in the space requirements, but lost in the intersection test. We also gained a little in
generalization: comparing the two algorithms yield that the second method is a little bit more general,
especially by the fact that we can add circles at arbitrary distance from the origin. We do not need this
for our DLA simulation, though.
Intersection handling
So now we have the data structure, but what do we do with the knowledge that two particles intersect?
Remember that we let it make discrete steps for simplification, but we are simulating a continuously
moving particle. A particle can therefore never step on another particle, but only touch a particle and
attach to it. The solution is simple: if we make a step and find an intersection, we move the particle
back into the direction where it came from until it only touches the other particle.
The above image shows how this position is calculated: we first draw a line L between the old position
and the new position (on which it intersects) of the particle. Then we draw a circle C that has twice the
diameter of an ordinary circle around the intersecting circle. We now place the particle on the
intersection of L and C.
�















 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc