Segmenting Internal Brain Nuclei in MRI Brain Image
using Morphological Operators
R.Dhanasekaran2
D.Selvaraj1
1Research Scholar, Department of Electronics &
Communication Engg., Sathyabama University
Chennai, India
1mails2selvaraj@yahoo.com
2Principal,
Syed Ammal Engineering College,
Ramanathapuram, India
2rdhanasekar@yahoo.com
Abstract— We present a new technique for segmenting brain
nuclei from MRI brain images. Our method performs the
segmentation using a combination approach of thresholding with
morphological operators. The MRI brain image contains skull
and noisy background. The latter have to be removed for further
analysis. Elimination of any obstacles and noise from the image is
the primary function of the morphological operators. We use
simple morphological operators like dilation, erosion, opening
and closing to the binarized MRI brain image. The results of
skull stripped MR image with the use of disk shaped structuring
elements are presented in the paper. The proposed method has
been applied to a large number of MR images showing promising
results for various image qualities, encouraging for future.
Keywords— Image segmentation, Image processing, skull
stripping, Morphological operator, brain segmentation
I. INTRODUCTION
Magnetic resonance imaging (MRI) of the human brain is
the most common type of medical imaging used in the
medical diagnosis among a variety of imaging modadilities
such as computer
tomography (CT), positron emission
tomography, ultrasound, mammography and radiography. So,
MR images are widely used not only for detecting tissue
deformities such as cancer and injuries but also for studying
brain pathology [8]. Also, many neurological diseases and
conditions alter the normal volume and regional distribution
of brain parenchyma (Gray and white matter), cerebrospinal
fluid. Such abnormalities are commonly related to the
conditions of hydrocephalus, cystic formation, brain atrophy
and tumour growth.
for
the brain
parenchyma, CSF volume and shape is segmentation. Image
segmentation is to divide the image into disjoint homogenous
regions, where all the pixels in the same class must have some
common characteristics but the major problems that affect the
quality of MRI segmentation are noise, inhomogeneous pixel
intensity distribution and blunt boundaries in the medical MR
images caused by MR data acquisition process [2, 3, and 4].
These problems do make manual quantitative analysis of brain
imaging data a tedious and time consuming procedure, prone
to observer variability [2]. Due to the characteristics of brain
MRI, development of automated segmentation algorithms
require pre-processing which includes denoising, stripping of
skull.
reliable measurement of
The basis
This paper presents a method for skull segmentation using a
sequence of mathematical morphological operations: erosion
and dilation, and their compositions i.e., opening and closing.
The operators of morphological processing are particularly
useful for the analysis of binary images so that MRI images
need to be previously binarized. The background and brain
mask of the image are obtained by applying a combination
approach of thresholding with morphology.
The next section presents some basics on morphological
operations. Section 3 describes our methodology. Finally we
show some results in section 4 and draw some conclusions
and future work perspectives in section 5.
II. MATHEMATICAL MORPHOLOGY CONCEPTS
that extracts
image objects
Mathematical morphology is a non-linear image analysis
technique
information by
describing its geometrical structure in a formal way [7].
Mathematical morphology has been largely used in several
practical image processing and analysis problems. Here we
briefly review some mathematical morphology operators and
the corresponding operations used in this work.
Mathematical operators take two data as an input: an image
to be processed and a structuring element, which is a matrix
used for defining a neighbourhood shape and size [1]. By
choosing the shape and size of the element, we can influence
the morphological operations sensitivity to specific shapes
appearing in the processed image. The elementary shapes of
symmetrical structuring elements used in the following
processing are shown in Fig. 1.
The erosion of binary image I by structuring element S is
The dilation of binary image I by structuring element S is
defined by the formula [1]:
(2)
I ⊕ S = {x,y : Sxy ∩ I≠∅}
Let f: D⊂ Rn → R is an image function and g: G⊂ Rn →
R is a structuring function. The two fundamental operations of
gray-scale morphology, erosion and dilation, are defined as:
Definition 1 (Dilation) The dilation of the function f(x) by
the structuring function g(x), (f⊕ g)(x), is given by:
(f ⊕ g)(x) = max {f(z) + (gx)(z) : z ∈ D[gx]} (3)
Definition 2 (Erosion) The erosion of the function f(x) by
the structuring function g(x), (f Θ g) (x), is given by:
(f Θ g)(x) = min {f (z) − (gx) (z): z ∈ D[gx]}
Where gx indicates the translation by x (gx) (z) = g(z − x),
(4)
and D[gx] denotes the domain of the translated structuring
function.
defined by the formula [1]:
I ⊗ S = {x,y : Sxy ⊆ I}
(1)
978-1-4244-5392-4/10/$26.00 ©2010 IEEE
�
The operations of closing and opening are the combinations
of erosion and dilation, both using the same structuring
element. Morphological opening is erosion followed by
dilation and morphological closing is dilation followed by
erosion. The Fig.3 shows that in a binarized image there are
some remaining pixels that represent the noise. To remove the
left-over pixels the opening operation was used.
(a)
(b)
Figure 1. Disk shape structuring elements: (a) 2-pixel radius, (b) 5-pixel
radius
III. PROPOSED METHODOLOGY FOR STRIPPING SKULL TO
SEGMENT BRAIN
This section presents
the proposed methodology for
segmenting brain MRI images. The fundamental task in brain
MRI segmentation is the classification of volumetric data into
grey matter, white matter and cerebrospinal fluid but it is not
easy as there are some inherent difficulties associated with
image segmentation; among them are RF coil in homogeneity,
brain tissue susceptibility and other systematic artifacts.
Various preprocessing steps have been proposed to deal with
some or all of these difficulties. Skull stripping is the first
processing step in the segmentation of brain tissueas shown in
Fig 2.
Figure 2. Overview of proposed methodology
In the proposed method for skull stripping, we see the brain
surface as a smooth manifold with relatively low curvature
that separates brain from non-brain regions. Also, the brain
cortex can be visualized as a distinct dark ring surrounding the
brain tissues in the T1-weighted axial MR images.
The steps involved in the proposed methodology for skull
stripping and brain extraction consists of three steps.
1. Binarization of every image.
2. Opening operation and closing operation on every image
in the sequence using the structuring element.
3. Applying the binary mask to the received MRI input
image.
A. Binarization
Binarization is the process that converts a grey level image
into a binary image. The binarization process involves
examining the grey-level value of each pixel in the enhanced
image, and if the value is greater than the global threshold,
then the pixel value is set to a binary value one; otherwise it is
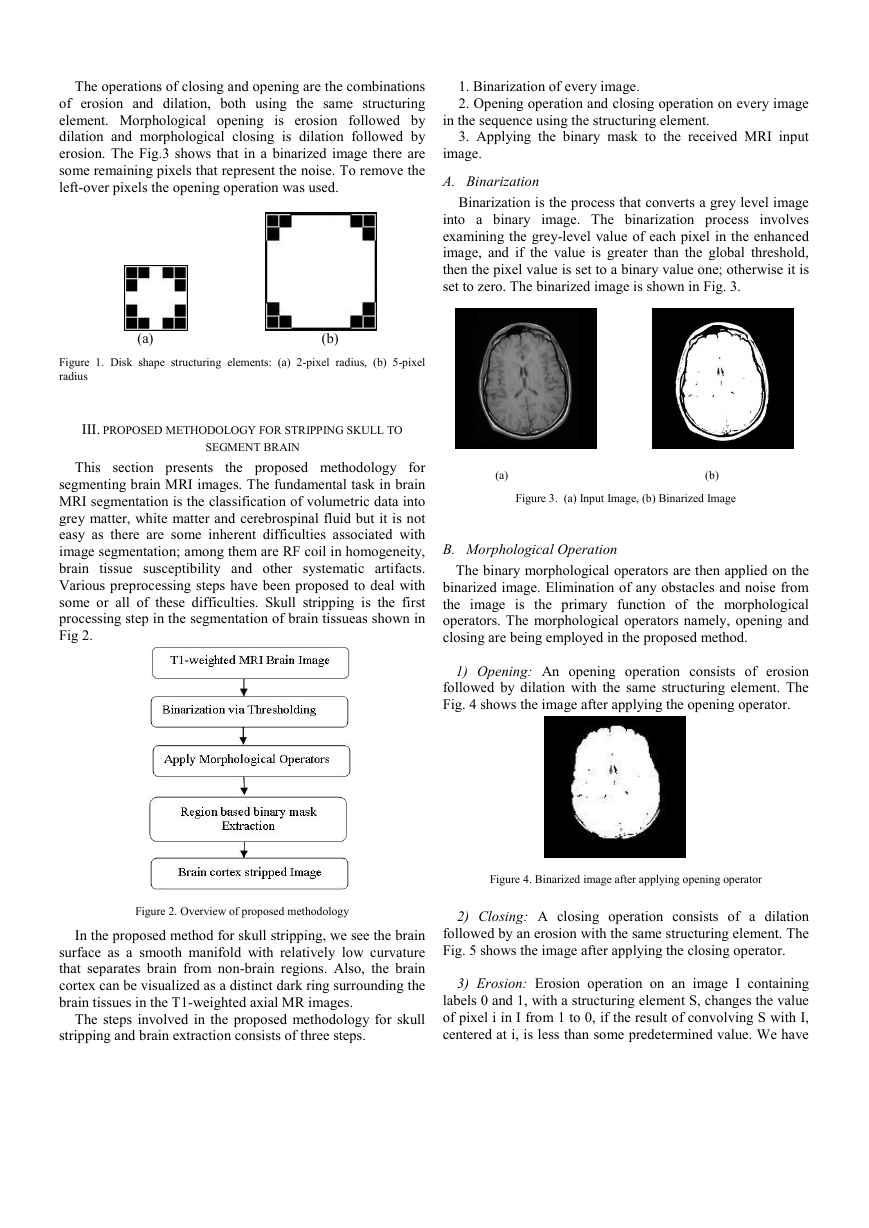
set to zero. The binarized image is shown in Fig. 3.
(a)
(b)
Figure 3. (a) Input Image, (b) Binarized Image
B. Morphological Operation
The binary morphological operators are then applied on the
binarized image. Elimination of any obstacles and noise from
the image is the primary function of the morphological
operators. The morphological operators namely, opening and
closing are being employed in the proposed method.
1) Opening: An opening operation consists of erosion
followed by dilation with the same structuring element. The
Fig. 4 shows the image after applying the opening operator.
Figure 4. Binarized image after applying opening operator
2) Closing: A closing operation consists of a dilation
followed by an erosion with the same structuring element. The
Fig. 5 shows the image after applying the closing operator.
3) Erosion: Erosion operation on an image I containing
labels 0 and 1, with a structuring element S, changes the value
of pixel i in I from 1 to 0, if the result of convolving S with I,
centered at i, is less than some predetermined value. We have
�
set this value to be the area of S, which is basically the
number of pixels that are 1 in the structuring element itself.
The structuring element (also known as the erosion kernel)
determines
thins
boundaries.
the details of how particular erosion
4) Dilation: Dual to erosion, a dilation operation on an
image I containing labels 0 and 1, with a structuring element S,
changes the value of pixel i in I from 0 to 1, if the result of
convolving S with I , centered at i , is more than some
predetermined value. We have set this value to be zero. The
structuring element (also known as the dilation kernel)
determines the details of how a particular dilation grows
boundaries in an image
Figure. 5. Brain Mask
C. Region-based binary mask extraction
Step 8: Develop binary mask choosing the larger area
Step 9: Apply binary mask on the original image to get
skull stripped image (brain nuclei).
IV. EXPERIMENTAL RESULTS
The experimental results of the proposed methodology for
segmenting brain MRI images are presented in this section.
The proposed methodology is implemented in Matlab (7.4).
The input to the proposed methodology is T1-weighted brain
MRI images collected from publicly available databases.
Regarding T1-weighting, every tissue in the human body has
its own T1 and T2 value. This term is used to indicate an
image where most of the contrast between tissues is due to
differences in the T1 value. The proposed methodology is
based on Intensity Thresholding (IT), which is the easiest and
fastest segmentation method, often adopted for preprocessing
of medical images and preregistration problems.
The sample results of brain MRI segmentation obtained
using the proposed methodology is shown in the following
Fig.7 to Fig. 11.
Region-based extraction
is done by examining
the
properties of each block that satisfy some criteria. We have
used one of two criteria. One criterion is to look at the max-
min difference and the other is to determine the mean values
of the blocks. The process results with a brain mask that is
then applied to the original MRI data. Consequently, we attain
a brain MRI image with its brain cortex stripped as shown in
Fig. 6.
(a) (b)
Figure 7. (a) Input Image, (b) Segmented Brain Image
Figure 6. Skull Stripped Brain Image
(a)
(b)
Figure 8. (a) Input Image, (b) Segmented Brain Image
D. Algorithm
Step 1: Get the MRI brain image to be stripped
Step 2: Find the maximum pixel value in the image
Step 3: Find the limit of high and Low frequency value
Step 4: Normalize the image
Step 5: Calculate the gray threshold from normalized
image
Step 6: Construct binary image using gray threshold value
Step 7: Apply disk shaped morphological operator to
eliminate the obstacle and noise from the image
(a) (b)
Figure 9. (a) Input Image, (b) Segmented Brain Image
�
(a) (b)
Figure 10. (a) Input Image, (b) Segmented Brain Image
(a)
Figure 11. (a) Input Image, (b) Segmented Brain Image
(b)
V. CONCLUSION
In this paper, an automated, simple and efficient brain MRI
segmentation method for classifying brain tissues has been
presented. Initially, the cortex present in the brain MRI
images is extracted by combining preprocessing techniques
and incorporating mathematical morphology. Experimental
results have showed that the proposed method does a
reasonably good job in terms of segmentation. In future from
this segmented brain image we can segment grey matter,
White matter and cerebrospinal fluid.
REFERENCES
[2]
[1] Renata Kalicka, Anna Pietrenko-Dabrowska, “Efficiency of new
method of removing the noisy background from the sequence of MRI
scans depending on structuring elements used to morphological
operations, Proceedings of the 2008 1st international conference on
information Technology, IT 2008, May 2008
Zijdenbos, A., Forghani, R., Evans, A.: “Automatic pipeline analysis of
3D MRI data for clinical trials: application to multiple sclerosis”, IEEE
transactions on medical imaging, Vol: 21, No: 10, pp: 1280–1291,
2002.
[3] Van-Leemput, K.: “Probabilistic brain atlas encoding using bayesian
inference”, Book Medical Image Computing and Computer-Assisted
Intervention (MICCAI), vol:1, pp:704–711, 2006.
[4] D. L. Pham, C. Xu, J. L. Prince, " A survey of current methods in
medical image segmentation", In Annual Review of Biomedical
Engineering, Vol. 2 , pp. 315-338, 2000.
Legal-Ayala, H.A. Facon, J., "Automatic segmentation of brain MRI
through learning by example", proceedings of International Conference
on Image Processing, (ICIP '04.), Vol.2, pp. 917- 920, 24-27 Oct. 2004.
Pham, D., Xu, C., Prince, J., ”Current methods in medical image
segmentation”, Annual Review of Biomedical Engineering Vol:2, pp:
315–337, 2000.
Leyza aldo Dorini, Rodrigo Minetto, “White Blood cell Segmentation
using morphological operators and scale-space analysis”A. Karnik,
“Performance of TCP congestion control with rate feedback:
TCP/ABR and rate adaptive TCP/IP,” IEEE computer Society, 294-
304, 2007.
[8] Atkins M.S., Mackiewich B.T., Fully automatic segmentation of the
brain in MRI, IEEE Transactions on Medical Imaging 17(1), 1998, pp.
98-107.
[7]
[5]
[6]
�







 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc