Analysing NMR Metabolomics data using
OPLS-DA
Background
A gene encoding MYB transcription factor, with unknown function, PttMYB76, was selected from a
library of poplar trees for metabolomic characterization of the growth process in Poplar trees.
Objective
The objective of this exercise is to shed some light on how PCA and OPLS-DA may be used in state-of-
the-art Metabolomics. In particular, the objectives are to:
• Demonstrate how to analyze metabolomics data from two sets of samples representing one
control group and one treated group
o Using PCA to review data, identify patterns and trends
• Demonstrate how to identify differences and putative biomarkers in different sample groups and
compare the strength of OPLS-DA compared to PCA
o Using OPLS-DA
• Describe the model diagnostics of an OPLS-DA model
Data
In total, the data set contains N = 57 observations, 6 trees divided into segments of 8 by the internode of
the tree plus analytical replicates and K = 655 variables (1H-NMR chemical shift regions bucket with
0.02ppm). The internode represents the growth direction of a plant. Internode 1 is the top of the plant and
8 is the bottom. The observations (trees) are divided in two groups (“classes”):
• MYB76 poplar plant (Ai, Bi, Ci)- Class 2
• Wild type Poplar plant (Di, Ei, Fi)- Class 1
The name settings A, B, C corresponds to MYB76 plants and D, E, F to the wild type (control) plants.
The i after the letter corresponds to the internode number of the plant. The last 12 experiments in the
dataset are analytical replicates i.e. samples that were run twice in the spectrometer. The analytical
replicates are marked with r1 or r2 after the internode number.
The plant material was analyzed by a 500 MHz NMR spectrometer equipped with a HR/MAS probe. The
1H NMR spectra were reduced by binning all of the data points over a 0.02 ppm region. Data points
between 4.2- 5.6 ppm, corresponding to water resonances, were excluded, leaving a total of 655 NMR
spectral regions as variables for the multivariate modelling. A more detailed description of the
experimental conditions is found in [1].
1 S. Wiklund et.al A new metabonomic strategy for analysing the growth process of the poplar tree. Plant Biotechnology Journal 2005 3 pp 353-362
SIMCA Tutorial
Analysing NMR Metabolomics data using OPLS-DA •••• 1
�
Import data
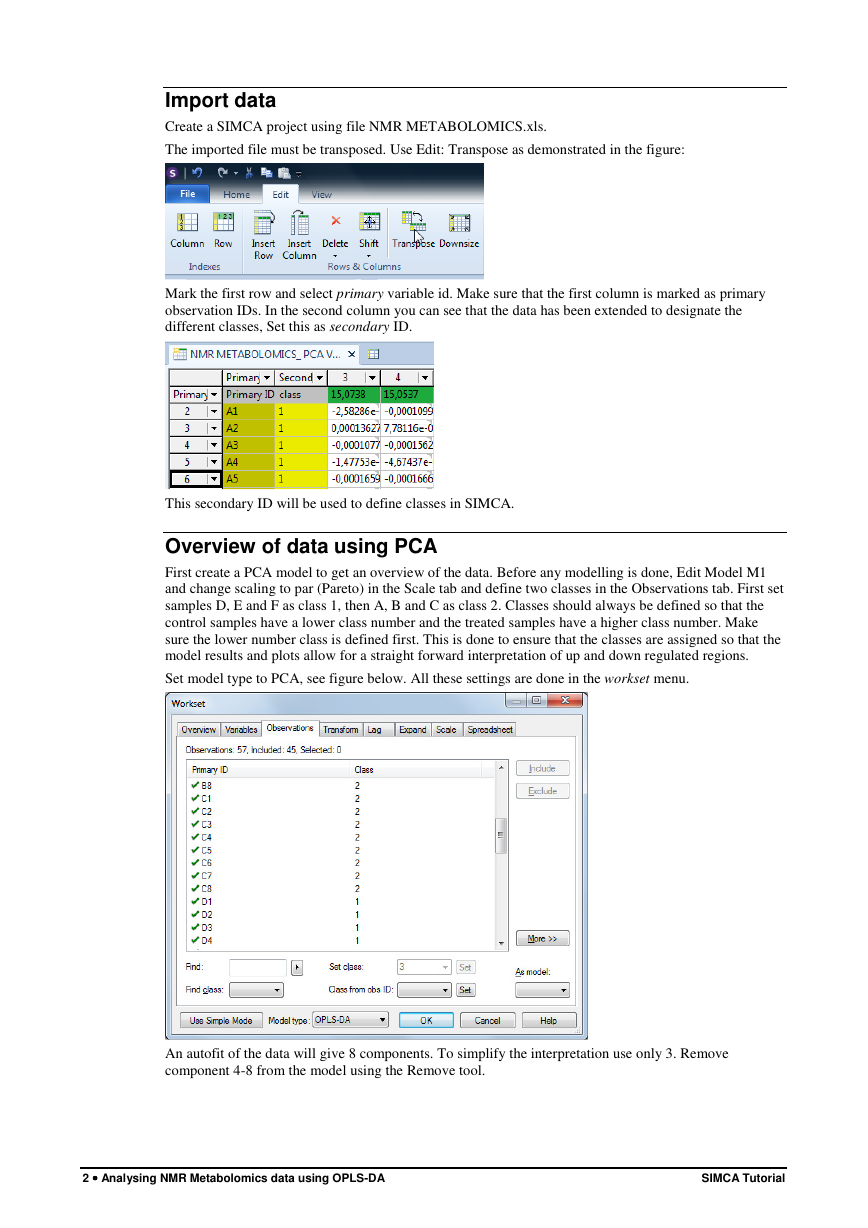
Create a SIMCA project using file NMR METABOLOMICS.xls.
The imported file must be transposed. Use Edit: Transpose as demonstrated in the figure:
Mark the first row and select primary variable id. Make sure that the first column is marked as primary
observation IDs. In the second column you can see that the data has been extended to designate the
different classes, Set this as secondary ID.
This secondary ID will be used to define classes in SIMCA.
Overview of data using PCA
First create a PCA model to get an overview of the data. Before any modelling is done, Edit Model M1
and change scaling to par (Pareto) in the Scale tab and define two classes in the Observations tab. First set
samples D, E and F as class 1, then A, B and C as class 2. Classes should always be defined so that the
control samples have a lower class number and the treated samples have a higher class number. Make
sure the lower number class is defined first. This is done to ensure that the classes are assigned so that the
model results and plots allow for a straight forward interpretation of up and down regulated regions.
Set model type to PCA, see figure below. All these settings are done in the workset menu.
An autofit of the data will give 8 components. To simplify the interpretation use only 3. Remove
component 4-8 from the model using the Remove tool.
2 •••• Analysing NMR Metabolomics data using OPLS-DA
SIMCA Tutorial
�
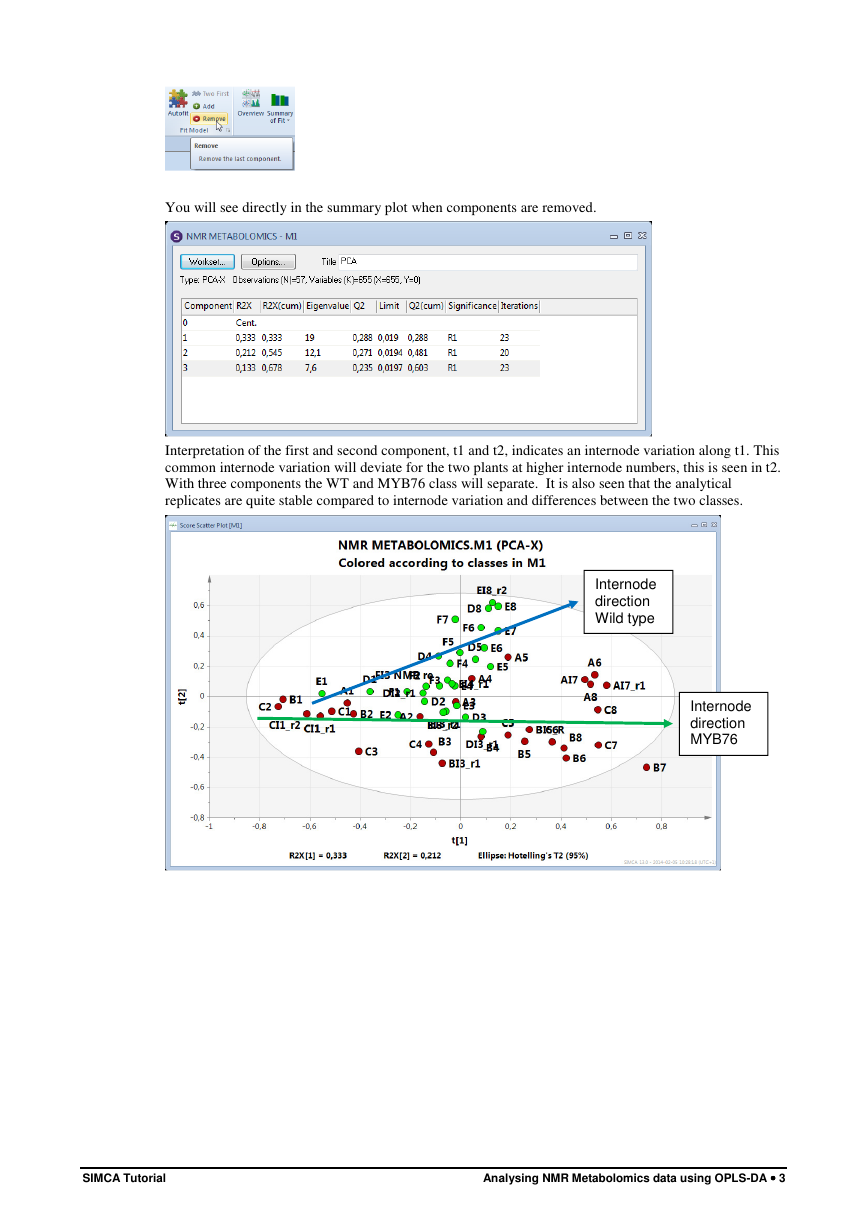
You will see directly in the summary plot when components are removed.
Interpretation of the first and second component, t1 and t2, indicates an internode variation along t1. This
common internode variation will deviate for the two plants at higher internode numbers, this is seen in t2.
With three components the WT and MYB76 class will separate. It is also seen that the analytical
replicates are quite stable compared to internode variation and differences between the two classes.
Internode
direction
Wild type
Internode
direction
MYB76
SIMCA Tutorial
Analysing NMR Metabolomics data using OPLS-DA •••• 3
�
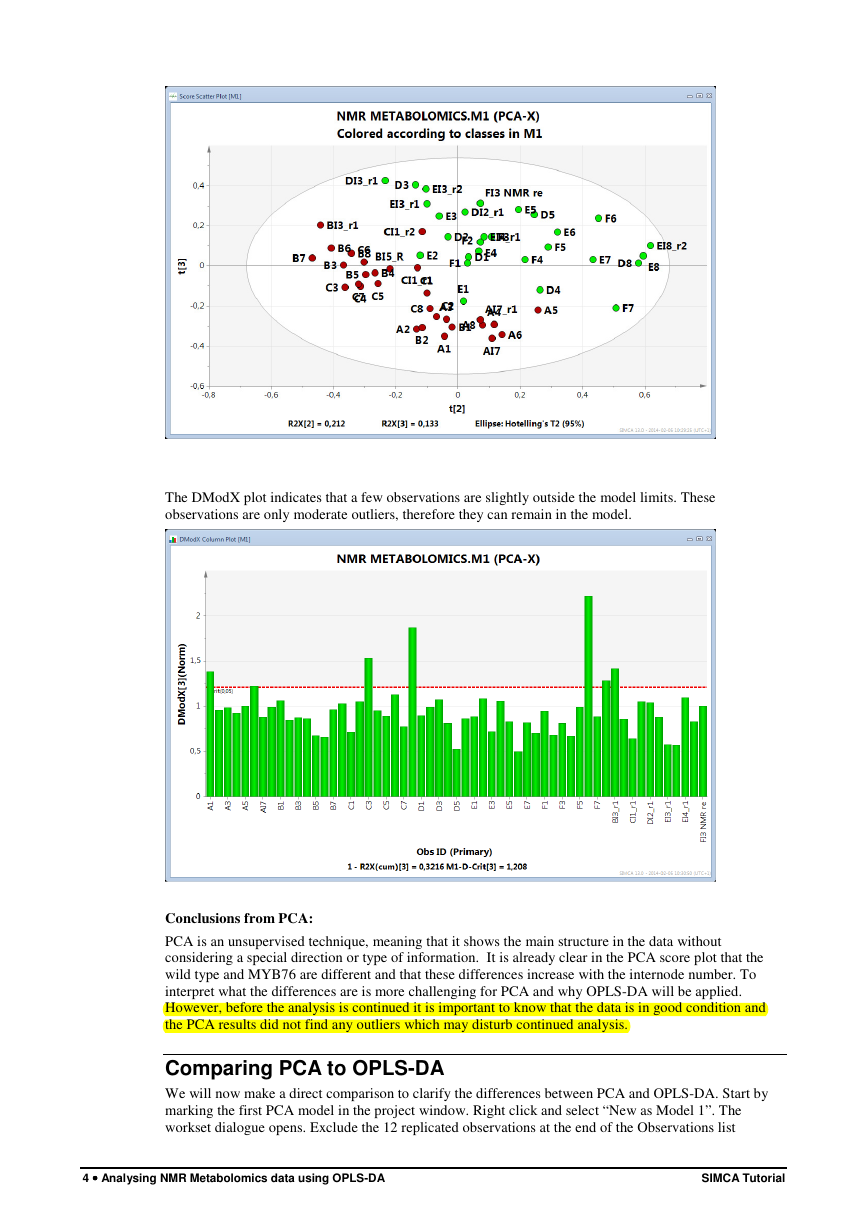
The DModX plot indicates that a few observations are slightly outside the model limits. These
observations are only moderate outliers, therefore they can remain in the model.
Conclusions from PCA:
PCA is an unsupervised technique, meaning that it shows the main structure in the data without
considering a special direction or type of information. It is already clear in the PCA score plot that the
wild type and MYB76 are different and that these differences increase with the internode number. To
interpret what the differences are is more challenging for PCA and why OPLS-DA will be applied.
However, before the analysis is continued it is important to know that the data is in good condition and
the PCA results did not find any outliers which may disturb continued analysis.
Comparing PCA to OPLS-DA
We will now make a direct comparison to clarify the differences between PCA and OPLS-DA. Start by
marking the first PCA model in the project window. Right click and select “New as Model 1”. The
workset dialogue opens. Exclude the 12 replicated observations at the end of the Observations list
4 •••• Analysing NMR Metabolomics data using OPLS-DA
SIMCA Tutorial
�
denoted r1 and r2. Model type should be PCA-X, press OK. Autofit the data and a 7 component model
appears.
The next step is to create an OPLS-DA model. Right click on model 2 and select “New as Model 2”.
Change the model type to OPLS-DA at the bottom of the Workset window.
Press OK. Autofit the model. A 1+4 OPLS model is created
Plot scores and compare the results from the PCA model to the OPLS-DA model. What can be seen in the
first OPLS-DA component? What can be seen in the orthogonal components?
A basic requirement for all prediction modeling is that the model is reasonably good with a high Q2.
Before doing any interpretation we need to check that the OPLS-DA model fulfils this requirement. In
this example we got a Q2 of 0,941 which is very high, so we can proceed to do model interpretation.
The advantage with the OPLS-DA model is that the between group variation (class separation) is seen in
the first component and within group variation will be seen in the orthogonal components. From the plots
below we see that the OPLS-DA model can be seen as a rotated PCA model.
PCA
OPLS-DA
NMR METABOLOMICS.M2 (PCA-X)
Colored according to classes in M2
NMR METABOLOMICS.M3 (OPLS-DA)
Scaled proportionally to R2X
Colored according to classes in M3
D3
B3
B6
B8
D2
E2
E3
E4
E5
F3
C3
B7
D1
C6
B4
F2
F1
C4
C7
B5
C5
B2
A2
C1
C2
E1
B1
A3
A1
C8
A8
D5
E6
F5
F4
D4
E7
A4 A5
A6
AI7
n
o
i
t
a
i
r
a
v
p
u
o
r
g
n
h
i
t
i
W
F6
E8
D8
F7
0,8
0,6
0,4
0,2
0
-0,2
-0,4
-0,6
-0,8
-1
E1
F7
D4
D8
E8
F1
E7
D1
F6
E2
E6
D2
D5
E5
F4F5
F3
F2
E4
E3
D3
C2
A1
C1
A2
A4
A5
AI7
B3
B4B5
C8
B6
B1
B2
C3
A3
C4
A8
A6
C5
C6
B8
C7
Between group variation
B7
-0,8
-0,6
-0,4
-0,2
0
0,2
0,4
0,6
1,00095 * t[1]
0,4
0,2
]
3
[
t
0
-0,2
-0,4
-0,6
-0,8
-0,6
-0,4
-0,2
0
0,2
0,4
0,6
R2X[2] = 0,22 R2X[3] = 0,112 Ellipse: Hotelling's T2 (95%)
t[2]
R2X[1] = 0,157 R2X[XSide Comp. 1] = 0,287 Ellipse: Hotelling's T2 (95%)
The difference between PCA and OPLS-DA is clearly visualized in the two plots above. In the PCA
model the difference between WT and MYB76 is seen in a combination of two components, 2 and 3. In
the OPLS-DA model the difference between WT and MYB76 is seen in the first component, 1. The
common internode variation is visualized in the scores from the second component, also called the first
orthogonal component, to1.
In short, the information that the PCA model distributes over component 2, 3 and so on is isolated in the
first OPLS-DA component. To have all between group differences isolated in one single component
simplifies interpretation and the identification of up and down regulated regions.
The simple reason why this is seen is because this is the nature of the OPLS algorithm. The algorithm will
rotate the plane and separate correlated variation (in this example the two classes) from uncorrelated
SIMCA Tutorial
Analysing NMR Metabolomics data using OPLS-DA •••• 5
�
variation between X and Y. Uncorrelated variation is also called orthogonal variation and is not related to
the observed response Y.
Technical Note: As OPLS rotates the first score vector t1 when additional components are computed the
t1 vs. to1 plot changes when you add additional components to the model. Make sure that the model is
optimized by using cross validation. Do NOT optimize the model by visualizing the class separation from
the score plot.
6 •••• Analysing NMR Metabolomics data using OPLS-DA
SIMCA Tutorial
�
Interpretation of OPLS-DA model
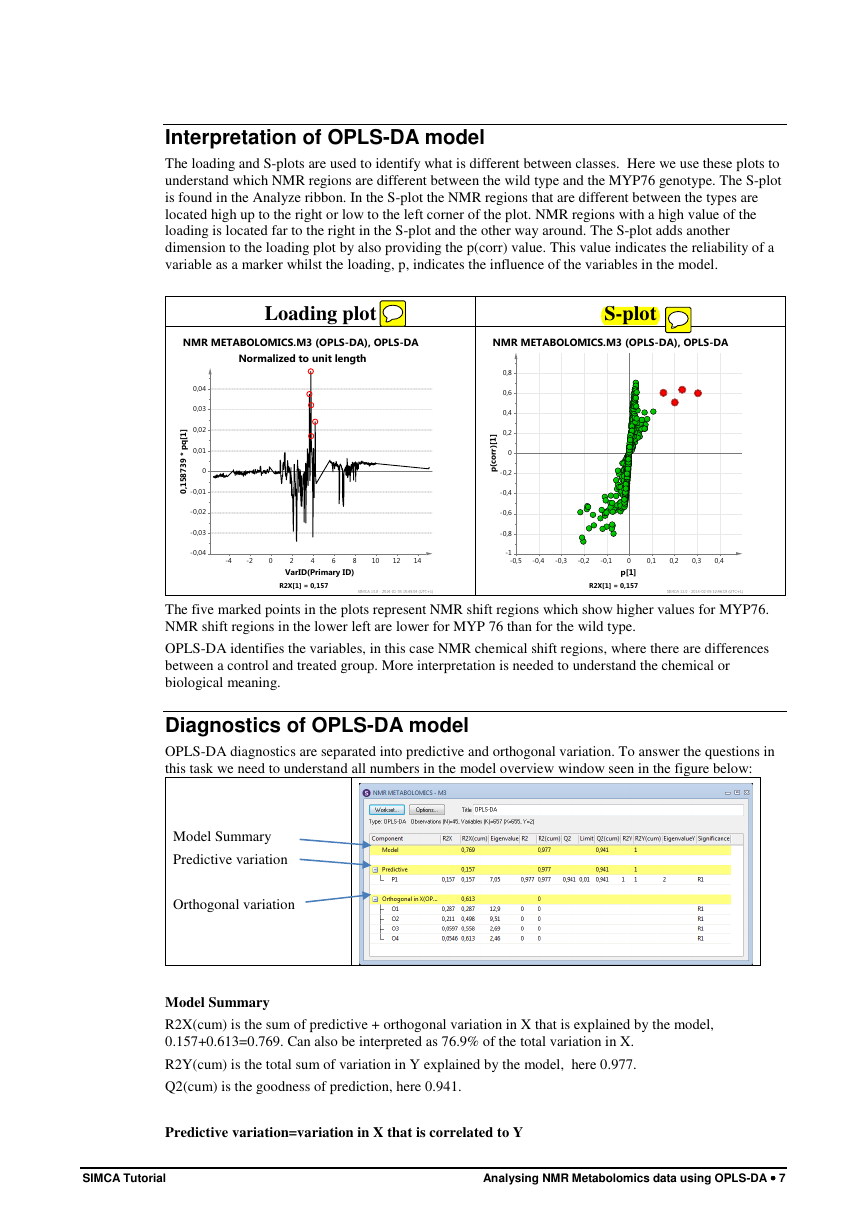
The loading and S-plots are used to identify what is different between classes. Here we use these plots to
understand which NMR regions are different between the wild type and the MYP76 genotype. The S-plot
is found in the Analyze ribbon. In the S-plot the NMR regions that are different between the types are
located high up to the right or low to the left corner of the plot. NMR regions with a high value of the
loading is located far to the right in the S-plot and the other way around. The S-plot adds another
dimension to the loading plot by also providing the p(corr) value. This value indicates the reliability of a
variable as a marker whilst the loading, p, indicates the influence of the variables in the model.
Loading plot
S-plot
NMR METABOLOMICS.M3 (OPLS-DA), OPLS-DA
NMR METABOLOMICS.M3 (OPLS-DA), OPLS-DA
Normalized to unit length
]
1
[
q
p
*
9
3
7
8
5
1
0
,
0,04
0,03
0,02
0,01
0
-0,01
-0,02
-0,03
-0,04
]
1
[
)
r
r
o
c
(
p
0,8
0,6
0,4
0,2
0
-0,2
-0,4
-0,6
-0,8
-1
-4
-2
0
2
4
6
8
10
12
14
-0,5
-0,4
-0,3
-0,2
-0,1
0
0,1
0,2
0,3
0,4
VarID(Primary ID)
R2X[1] = 0,157
p[1]
R2X[1] = 0,157
The five marked points in the plots represent NMR shift regions which show higher values for MYP76.
NMR shift regions in the lower left are lower for MYP 76 than for the wild type.
OPLS-DA identifies the variables, in this case NMR chemical shift regions, where there are differences
between a control and treated group. More interpretation is needed to understand the chemical or
biological meaning.
Diagnostics of OPLS-DA model
OPLS-DA diagnostics are separated into predictive and orthogonal variation. To answer the questions in
this task we need to understand all numbers in the model overview window seen in the figure below:
Model Summary
Predictive variation
Orthogonal variation
Model Summary
R2X(cum) is the sum of predictive + orthogonal variation in X that is explained by the model,
0.157+0.613=0.769. Can also be interpreted as 76.9% of the total variation in X.
R2Y(cum) is the total sum of variation in Y explained by the model, here 0.977.
Q2(cum) is the goodness of prediction, here 0.941.
Predictive variation=variation in X that is correlated to Y
SIMCA Tutorial
Analysing NMR Metabolomics data using OPLS-DA •••• 7
�
A corresponds to the number of correlated components between X and Y. If only one response vector is
used then A is always 1.
R2X is the amount of variation in X that is correlated to Y, here 0.157.
Orthogonal variation=variation in X that is uncorrelated to Y
A corresponds here to the number of uncorrelated (orthogonal) components. Each orthogonal component
is represented and can be interpreted individually.
R2X is the amount of variation in X that is uncorrelated to Y. Each component is represented
individually.
R2X(cum) in bold is the total sum of variation in X that is uncorrelated to Y, here 0.613.
Noise=1- 0.157 – 0.613=0.23 � 23%
Conclusions
• OPLS-DA is an excellent tool for “omics” data analysis due to its ability to pinpoint differences
between groups of observations and disregard disturbing structure in data.
• OPLS-DA is the discriminant version of OPLS
• OPLS separates correlated (predictive) variation from uncorrelated (orthogonal) variation
between X and Y.
•
In OPLS-DA studies with two groups, the predictive component, t1, will describe the differences
between two groups and the orthogonal components will describe systematic variation in the
data that is not correlated to Y.
• The separation of predictive and orthogonal components will facilitate interpretation of
metabolomics data in terms of model diagnostics and also for biomarker identification. The later
will be described in another example.
• OPLS in Metabolomics studies allows the user to mine complex data and provides information
which allows us to propose intelligent hypotheses.
8 •••• Analysing NMR Metabolomics data using OPLS-DA
SIMCA Tutorial
�












 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc