Journal of Neuroscience Methods 223 (2014) 50– 68
Contents lists available at ScienceDirect
Journal of Neuroscience Methods
j o u r n a l h o m e p a g e : w w w . e l s e v i e r . c o m / l o c a t e / j n e u m e t h
Computational Neuroscience
The MVGC multivariate Granger causality toolbox: A new approach to
Granger-causal inference
Lionel Barnett∗, Anil K. Seth
Sackler Centre for Consciousness Science and School of Informatics, University of Sussex, Brighton BN1 9QJ, UK
h i g h l i g h t s
• Matlab© Toolbox for accurate and efficient calculation of Granger causality.
• Calculate Granger causalities (conditional and unconditional) in both time and frequency domains.
• Based on advanced VAR (vector autoregression) theory.
• Avoids separate “full” and “reduced” regressions.
a r t i c l e
i n f o
a b s t r a c t
Article history:
Received 5 September 2013
Received in revised form 16 October 2013
Accepted 26 October 2013
Keywords:
Granger causality
Vector autoregressive modelling
Time series analysis
1. Introduction
causality
(G-causality)
Wiener–Granger
(Granger, 1969;
Geweke, 1982, 1984) is an increasingly popular method for identi-
fying “causal” connectivity in neural time series data (Bressler and
Seth, 2011). It can be traced conceptually to Wiener (1956) and
was operationalised by Granger in terms of linear autoregressive
∗ Corresponding author. Tel.: +44 1273 678101.
E-mail addresses: l.c.barnett@sussex.ac.uk (L. Barnett), a.k.seth@sussex.ac.uk
(A.K. Seth).
0165-0270/$ – see front matter ©
http://dx.doi.org/10.1016/j.jneumeth.2013.10.018
2013 Elsevier B.V. All rights reserved.
Background: Wiener–Granger causality (“G-causality”) is a statistical notion of causality applicable to time
series data, whereby cause precedes, and helps predict, effect. It is defined in both time and frequency
domains, and allows for the conditioning out of common causal influences. Originally developed in the
context of econometric theory, it has since achieved broad application in the neurosciences and beyond.
Prediction in the G-causality formalism is based on VAR (vector autoregressive) modelling.
New method: The MVGC Matlab© Toolbox approach to G-causal inference is based on multiple equiva-
lent representations of a VAR model by (i) regression parameters, (ii) the autocovariance sequence and
(iii) the cross-power spectral density of the underlying process. It features a variety of algorithms for
moving between these representations, enabling selection of the most suitable algorithms with regard
to computational efficiency and numerical accuracy.
Results: In this paper we explain the theoretical basis, computational strategy and application to empirical
G-causal inference of the MVGC Toolbox. We also show via numerical simulations the advantages of our
Toolbox over previous methods in terms of computational accuracy and statistical inference.
Comparison with existing method(s): The standard method of computing G-causality involves estimation
of parameters for both a full and a nested (reduced) VAR model. The MVGC approach, by contrast, avoids
explicit estimation of the reduced model, thus eliminating a source of estimation error and improving
statistical power, and in addition facilitates fast and accurate estimation of the computationally awkward
case of conditional G-causality in the frequency domain.
Conclusions: The MVGC Toolbox implements a flexible, powerful and efficient approach to G-causal
inference.
© 2013 Elsevier B.V. All rights reserved.
modelling of stochastic processes (Granger, 1969). G-causality is
based on predictability and precedence. Put simply, a variable X is
said to G-cause a variable Y if the past of X contains information that
helps predict the future of Y over and above information already
in the past of Y. Importantly, G-causality is a measure of directed
functional connectivity in terms of providing a statistical descrip-
tion of observed responses. In contrast, methods for identifying
effective connectivity aim to elucidate the “the simplest possible
circuit diagram explaining observed responses” (Valdes-Sosa et al.,
2011; Aertsen and Preißl, 1991; Friston et al., 2003, 2013).
The MVGC Matlab Toolbox implements numerical routines for
calculating multivariate Granger causality (MVGC) from time series
�
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
51
data, both unconditional and conditional, in the time and frequency
domains. It supersedes and extends the GCCA (Granger Causal Con-
nectivity Analysis) Toolbox (Seth, 2010). Based on advanced VAR
(vector autoregressive) model theory (Hamilton, 1994; Lütkepohl,
2005), it is designed for computational efficiency and accuracy.
Improving upon standard approaches to G-causal inference, it
avoids separate full and reduced regressions (see Section 2),
thus eliminating a common source of statistical inaccuracy and
computational inefficiency. It also facilitates estimation of fully
multivariate conditional G-causality in the frequency domain.
The MVGC toolbox has been designed with application to empir-
ical neuroscience data in mind. Several issues commonly faced in
applying G-causal inference to such data are discussed later in the
paper (Section 4). However, the software is not restricted to this
application domain. G-causal inference is a very general method
which has been productively applied in many areas, though caution
is always needed in ensuring that the data satisfy the assump-
tions underpinning the method (see Section 2.1 and also Section
3.3). Thus, while the MVGC software is designed to be intuitive
and straightforward to use, to get the most out of it some under-
standing of the theoretical basis of G-causal inference, as well
as approaches to its numerical computation, is recommended. To
facilitate this, this paper presents the conceptual and theoretical
basis of G-causality (substantially extending previous presenta-
tions), and details the computational implementation of the MVGC
toolbox. Although not intended as a user guide—the integrated
help system and walk-through demonstration scripts fulfill that
purpose—Section 3 provides a helpful overview of the toolbox
rationale and design principles, Section 3.1 explains the MVGC
computational strategy while Appendices A.1 and A.2 detail the
core computational algorithms.
2. G-causality: theory, estimation and inference
Y
Y
Y
X
X
=
Y 1,
X1,
X2, . . .,
Y; intuitively, past values of
X
=
X
if and only if
does convey information about the future of
Assume two jointly distributed vector-valued stochastic pro-
Y 2, . . .. We say that
cesses (“variables”)
Y
X, conditional on its own past,
does not G-cause
yield
is independent of the past of
no information about the current value of
beyond information
itself. If, conversely, the past
already contained in the past of
of
above and
then we say that
beyond all information contained in the past of
Y
X. Much has been made of the fact that this notion of
“causality” does not necessarily tally with more conventional (and
perhaps more physically intuitive) notions (Pearl, 2009; Valdes-
Sosa et al., 2011; Friston, 2011; Roebroeck et al., 2009, 2010) (note,
for example, that the processes must be strictly non-deterministic
for the above definition even to make sense). We do not engage
this debate here; throughout this paper the term “causal” is used
exclusively in the Wiener–Granger sense just described.
G-causes
X
X
Testing for Granger (non-)causality requires establishing—in
a statistical sense—a conditional (non-)dependency. This may
seem to invite an information-theoretic approach, since mutual
information is a natural measure of statistical dependence. How-
ever in empirical settings a drawback with information-theoretic
quantities is the difficulty of estimation in sample and lack of
known theoretical distributions for information-theoretic esti-
mators, complicating statistical inference (but see Barnett and
Bossomaier, 2013). An alternative is therefore to invoke a model-
based or parametric approach for which conditional dependency
estimators are more efficient and (preferably) have known samp-
ling distributions, thus facilitating inference. In fact, G-causality is
frequently identified with a model-based viewpoint, which may
also yield intuitive predictive interpretations of G-causality; for
X, conditional on its own past,
predictive models, the statement “
is independent of the past of
X
Y
predicted by its own past”.
does not help predict
Y” may be reframed as “the past of
may be
beyond the degree to which
X
The most common operationalisation of G-causality, and the one
on which the MVGC Toolbox is based, utilises VAR modelling of time
um, where for
series data. A multivariate time series
ut is a real-valued n-dimensional (column) vector with
each time t
components u1t, u2t, . . ., unt, is considered as a realisation of length
m of a discrete-time stationary1 vector stochastic process
U2,
. . . (the “universe of variables”). A pth order vector autoregressive
model for the process—a VAR(p)—takes the form2
u2, . . .,
U1,
u1,
U t =
Ak ·
U t−k +
εt
(1)
p
k=1
×
Here p is the model order, which may be infinite. The n
n
real-valued matrices Ak are the regression coefficients, and the
εt the residuals, which are
n-dimensional stochastic process
independently and identically distributed (iid) and serially uncor-
is, they constitute a white noise process. The
related; that
×
parameters of the model are the coefficients Ak and the n
n resid-
≡
cov(εt) which, by stationarity, does not
uals covariance matrix �
depend on the time t. Interpreted as a predictive model, (1) thus
models the value of the process at current time t in terms of its
−
past values at times t
p. The regression coefficients
represent the predictable structure of the data, the residuals the
unpredictable.
−
1, . . ., t
The determination of a VAR model (1) should not be taken to
imply that the time series data modelled by the stochastic pro-
ut was actually generated by a linear autoregressive scheme.
cess
In comparison to effective connectivity techniques like dynamic
causal modelling [DCM] (Friston et al., 2003), which make very
explicit assumptions about the generative mechanism underlying
the observed data (Friston et al., 2013), the VAR models underlying
G-causality are “generic”, in the sense that they make no assump-
tions about the mechanism that produced the data, beyond that a
VAR model actually exists. Standard theory (Anderson, 1971) yields
that a rather general class of covariance-stationary multivariate
process—including many nonlinear processes—may be modelled
as VARs, albeit of theoretically infinite order (see also the next
section). Note that this belies a common misapprehension that
VAR-based G-causal analysis is restricted to linear processes. In
practice, given empirical time-series data, a finite model order p
is selected on theoretical principles, sufficiently high so as to cap-
ture predictable variation but not so high as to overfit unpredictable
variation in the data (Section 2.4).
2.1. VAR process theory
The MVGC toolbox exploits some advanced aspects of VAR pro-
cess theory, which we outline here.
For valid G-causality analysis, the VAR coefficients in (1) must
be square summable and stable (Hamilton, 1994; Lütkepohl, 2005;
∞
Rozanov, 1967). Square summability requires that
Ak2 <
p
k=1
1 G-causal inference is not necessarily limited to stationary processes. Under some
circumstances—notably for multi-trial data or for near-stationary data that may be
“windowed”—the stationarity requirement may be relaxed. This is addressed later
on.
2 Throughout, bold symbols represent vector quantities and normal typeface
represents scalar quantities. Capitalised symbols represent random variables or
U t for the random vector representing
matrices, according to context. Thus we write
U 2, . . . at time t,
um for a realisation of the pro-
u1, . . .,
the value of the process
ut, etc. A superscript
cess of length m, uit for the ith component of
denotes matrix
∗
transpose, and a superscript
denotes (complex) matrix conjugate-transpose. |M|
denotes the determinant of the square matrix M.
U 1,
�
52
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
For a VAR process, the CPSD admits a unique spectral factorisation
(Papoulis and Pillai, 2002)
≤
∗
=
H()�H()
S()
0
≤
2
(9)
where the transfer function H() is defined as the inverse matrix of
the Fourier transform of the regression coefficients:
(2)
≡
H()
−
I
−ik
Ake
≤
≤
2
0
(10)
−1
p
k=1
which means intuitively that the coefficients do not “blow up” even
for infinite model order p (for finite p square summability is sat-
isfied trivially). Stability means that the VAR coefficients define a
covariance-stationary process. This is related to the the characteristic
polynomial for the coefficients sequence Ak, which is:
I
p
Akzk
k=1
≡
ϕA(z)
−
for the variable z defined in the complex plane
C. By standard the-
ory (Lütkepohl, 2005) the coefficients Ak define a stable VAR iff the
≤
characteristic polynomial is invertible on the unit disc |z|
1 in the
complex plane. Defining the spectral radius of the VAR as
(A)
≡
max
ϕA(z)=0
{|z|−1}
(3)
an equivalent condition
covariance-stationary process—is
for stability—i.e.
that
(1) define a
(A) < 1
(4)
−2,
cov(ut,
=
. . .,
ut−k) k
−1, 0, 1, 2, . . .
The autocovariance sequence �k for a covariance-stationary (not
ut is defined as the sequence
necessarily VAR) stochastic process
×
of n
n matrices
�k ≡
Note that by stationarity the �k do not depend on the time t, and
for all lags k. For a VAR process of the form (1), it may
that �−k = �k
be shown [Appendix A.1, (A5)] that in the long run, the autocovari-
ance sequence decays exponentially with lag k, the rate of decay
being just the spectral radius (A). Note, importantly, that this
implies that if the autocovariance sequence for a process does not
decay (sub-)exponentially then the process may not be modelled
as a VAR (see Section 3.3). This has some importance for analysis of
neural data, which we return to in Section 4.2.
(5)
For a VAR process (1), the autocovariance sequence is related
to the VAR parameters via the Yule-Walker equations (Anderson,
1971)
�k =
A�k− +
ık0� k
=
. . .,
−2,
−1, 0, 1, 2, . . .
(6)
p
=1
Numerically, there are efficient algorithms available for solving (6)
for the �k in terms of (Ak, �) [Appendix A.1, (A5)] and, conversely,
for (Ak, �) in terms of the �k [Appendix A.1, (A6)].
The cross-power spectral density (CPSD) is defined as the (two-
sided) Fourier transform of the autocovariance sequence3:
=
S()
−ik 0
≤
≤
2
(7)
∞
�ke
k=−∞
×
The S() are n
n Hermitian positive semi-definite matrices. The
inverse Fourier transform recovers the autocovariance sequence:
�k = 1
2
−1, 0, 1, 2, . . .
S()eik d k
=
. . .,
−2,
(8)
−
Numerically, (7) and (8) may be efficiently computed by a (discrete)
Fast Fourier Transform (FFT) and Inverse Fast Fourier Transform
(IFFT) respectively.
While there is no known closed-form solution of (9) for H(), � in
terms of S(), a classical result (Masani, 1966; Wilson, 1972) states
that, given a CPSD S() of a VAR process, a unique solution exists4
and may be computed numerically [Appendix A.1, (A7)]. According
to (10) the VAR coefficients Ak may then be recovered from H() by
a matrix inversion and inverse Fourier transform.
A further technical condition for valid G-causality analysis is that
the CPSD be uniformly bounded away from zero almost everywhere
(Geweke, 1982, 1984). Explicitly, there must exist a c > 0 such that
for almost all 0
c−1I ≤
≤
cI
≤
2
S
≤
(11)
(λ)
−
where for A, B square matrices,
A is pos-
itive semidefinite. Importantly, condition (11) guarantees square
summability of the regression coefficients (Geweke, 1982, 1984;
Rozanov, 1967).
B denotes that B
≤A
Eqs. (6)–(10) relate the VAR parameters (Ak, �), the autoco-
variance sequence �k and the CPSD S(). Importantly, these three
mathematical objects specify entirely equivalent representations for
a VAR and any of them may be estimated from empirical time series
data. As we shall see (Section 3), the MVGC toolbox makes extensive
use of these equivalences to furnish accurate and efficient compu-
tational pathways for the calculation of G-causality in the time and
frequency domains.
2.2. Unconditional G-causality in the time domain
In its simplest (unconditional, time-domain) form, G-causality
Ut is split into two jointly
is motivated as follows: suppose that
distributed (i.e. inter-dependent) multivariate processes:
U t =
Xt
Y t
Y
Under a predictive interpretation (cf. Section 2), the G-causality
FY→X, stands to quantify the “degree to which
from
to
X, over and above the degree to which
the past of
X
is already predicted by its own past”. In the VAR formulation, this
notion is operationalised as follows: the VAR(p) (1) decomposes as
X, written
Y
helps predict
Xt
Y t
=
p
k=1
and the residuals covariance matrix as
≡
cov
�
x,t
εy,t
=
�xx �xy
�yx �yy
Axx,k Axy,k
Ayx,k Ayy,k
Xt−k
Y t−k
+
εx,t
εy,t
(12)
(13)
(14)
≤
3 For simplicity, all spectral quantities are defined on the normalised fre-
≤
2. In an empirical scenario—and indeed in the MVGC
quency range 0
≤
implementation—the natural frequency range of definition is 0
f where f is the
sampling frequency in Hertz (Hz). In fact, to take advantage of inherent symmetries,
the MVGC computes all spectral entities only up to the Nyqvist frequency f/2.
≤
4 More precisely, given S() there is a unique holomorphic matrix function ˜H(z) on
=
the complex plane with ˜H(0)
I, and a unique symmetric positive definite matrix
≡ ˜H(e
�, such that (9) holds for H()
−i).
�
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
53
p
k=1
p
k=1
p
k=1
The x-component of the regression (13) is
Xt =
Axx,k ·
Xt−k +
Axy,k ·
Y t−k +
εx,t
(15)
Y,
from which we see that the dependence of
given its own past, is encapsulated in the coefficients Axy,k; in par-
ticular, there is no conditional dependence of
on the past of
·
Y
= Axy,p = 0. This leads to consideration of the
iff Axy,1 = Axy,2 =
reduced (or restricted) regression—as opposed to the full regression
(15)—given by omitting the “historic” (past)
on the past of
dependency:
X
X
Y
·
·
Xt =
A
xx,k
·
Xt−k +
ε
x,t
(16)
X
so that now
xx,k are
is predicted by its own past only. Here A
ε
the reduced regression coefficients and
x,t the reduced regres-
≡
FY→X, then,
cov(
sion residuals, with covariance matrix �
xx
stands to quantify the degree to which the full regression (15) rep-
resents a “better” model of the data than the restricted regression
(16). Maximum likelihood (ML) theory (Edwards, 1992) furnishes a
natural framework for the analysis of parametric data modelling.
In this framework, an appropriate comparative measure for nested
models of the form (15) and (16) is the likelihood ratio statistic or,
for its more convenient statistical properties, its logarithm.5 Tech-
nically, it is a test statistic for the null hypothesis of zero causality
x,t).
·
·
·
(17)
Axy,2 =
=
Axy,p =
0
H0 : Axy,1 =
in (15). This motivates the definition of the G-causality statistic as
the appropriate log-likelihood ratio. Now standard ML theory tells
us that the likelihood for a realisation of length m of a VAR model
of the form (1) is proportional to |�|−(m−p)/2, where |�|, the gener-
alised variance of the model (Barrett et al., 2010), is the determinant
of the residuals covariance matrix. Thus the (unconditional, time-
domain) G-causality from
is defined to be the log-likelihood
ratio
FY→X ≡
to
(18)
ln
X
Y
|�
|
xx
|�xx|
=
cov(
where �xx = cov(εx,t) and �
x,t) are the residuals covari-
xx
ance matrices of the VAR models (15) and (16) respectively. A
predictive interpretation of (18) is that the generalised variance
of a regression model may be viewed as a measure of model predic-
tion error.6 From this perspective, G-causality (18) thus quantifies
the reduction in prediction error when the past of the process
is
included in the explanatory variables of a VAR model for
X.
Y
An important and frequently misunderstood point is that G-
causality is often perceived to lack quantitative meaning beyond
its interpretation as a hypothesis-test statistic appropriate only
for significance testing (Section 2.5); that is, that its magnitude is
essentially meaningless, and in particular should not be compared
with other G-causality statistics. However, G-causality magnitudes
5 Alternative test statistics which appear in the G-causality literature are the Wald
or Lagrange multiplier estimators (Edwards, 1992). An advantage of the likelihood
ratio is that is has pleasant invariance properties under a broad class of transforma-
tions (Barrett et al., 2010; Barnett and Seth, 2011) (Section 2.7), a natural spectral
decomposition (Geweke, 1982) (Section 2.6) and an information-theoretic interpre-
tation as a transfer entropy (Schreiber, 2000; Kaiser and Schreiber, 2002; Barnett
et al., 2009; Barnett and Bossomaier, 2013).
6 It may seem intuitive to use the mean square error trace(�) (the “total error”)
as a measure of prediction error. However, on the grounds of transformation invari-
ance, frequency decomposition and information-theoretic interpretation, as well as
for consistency with the ML formulation, the generalised variance is more appro-
priate. See Barrett et al. (2010) for a fuller discussion.
have a natural interpretation in terms of information-theoretic bits-
per-unit-time. This is because, for a large class of joint processes,
G-causality and information-theoretic transfer entropy (Schreiber,
2000) are asymptotically equivalent (Barnett and Bossomaier,
2013) [in the Gaussian case the equivalence is exact (Barnett et al.,
2009)]. Note that transfer entropy is conceptually isomorphic to G-
causality and is often described as a measure of information flow
(but see e.g. Lizier and Prokopenko, 2010). Thus, G-causalities may
be meaningfully compared, and magnitudes cited, albeit with due
caution to statistical significance (Section 2.5).
2.3. Conditional and pairwise-conditional G-causality
Y
X
X
and
and a third set of variables,
The unconditional G-causality statistic introduced above has the
undesirable characteristic that if there are joint (possibly histori-
Z
cal) dependencies between
say, then spurious causalities may be reported. Thus, for instance,
if there is no direct causal influence
but there are (possibly
Y
X
lagged) dependencies of
causality may be reported. These spurious causalities may be elimi-
nated by “conditioning out” the common dependencies – provided
they are available in the data. If, however, there are dependencies
on unknown (exogenous) or unrecorded (latent) variables, then
it will in general be impossible to eliminate entirely their poten-
tially confounding effect on causal inference, although attempts
have been made to mitigate their impact [e.g. “partial” G-causality
(Guo et al., 2008)7].
→
X
Z
then a spurious
Y
on
and
→
Y
To illustrate the conditional case, suppose that the universe
of
known, recorded variables splits into three jointly distributed (i.e.
inter-dependent) multivariate processes
U
⎞⎟⎠
⎛⎜⎝ Xt
Y t
Zt
U t =
(19)
and we wish to eliminate any joint effect of
the G-causality from
to (13) and we may consider the full and reduced regressions
Z
on the inference of
X. Again the VAR(p) (1) splits analogously
to
Y
p
p
k=1
k=1
Xt =
Xt =
Axx,k ·
Xt−k +
A
xx,k
·
Xt−k +
p
p
k=1
k=1
p
k=1
Axy,k ·
Y t−k +
Axz,k ·
Zt−k +
εx,t
(20)
A
xz,k
·
Zt−k +
ε
x,t
(21)
Zt
analogous to (15) and (16), but with the conditioning variables
included in both regressions. The null hypothesis to be tested is still
→
Z, which we write
Y
(17) and the causality
FY→X|Z, is again as in (18):
FY→X|Z ≡
conditioned on
(22)
ln
X
|�
|
xx
|�xx|
Z
in both regressions accounts for its joint
FY→X|Z may thus be read as “the degree to which the past of
is already
but now the inclusion of
effect.
Y
predicted by its own past and the past of
X, over and above the degree to which
helps predict
Z”.
X
Note that in this (and the previous) section, the source, target
Z
and conditioning variables
may themselves be multi-
variate; that is, they may represent groups of variables. It is in this
X,
Y,
7 Note that there are a number of errors in this paper: firstly, it is stated
erroneously that partial G-causality may be negative. Secondly, the sampling distri-
bution given is incorrect. See Roelstraete and Rosseel (2012) and also Barrett et al.
(2010).
�
54
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
sense that we use the term “multivariate” G-causality. G-causality
is thus able to account for group interactions. This is significant,
since elements in a multivariate system may function cooperatively
or competitively, or interact generally in a more complex fashion
than traditional bivariate analysis can accommodate (Ladroue et al.,
2009; Barrett et al., 2010).
U
A case of particular importance is that of pairwise-conditional
G-causality. Given a universe
of variables comprising n (known,
recorded) jointly distributed univariate processes U1t, . . ., Unt, it
is frequently of interest to estimate the G-causalities between
/=
pairs of variables Ui, Uj, i
j. The traditional bivariate pairwise
FUj
→Ui . Since these are prone to spurious
G-causalities are the
effects through joint dependencies as described above, however,
it is generally preferable to consider rather the pairwise-conditional
causalities
FUj
Gi,j(U)
|U[ij]
→Ui
(23)
≡
where the subscript [ij] denotes omission of the ith and jth variables
U. Thus, when considering the causal-
in the multivariate universe
ity Uj →
Ui the joint dependencies of all remaining known variables
/=
are conditioned out.8 The quantities
j may be considered
as a weighted directed graph, which we shall sometimes refer to as
the (G-)causal graph.
Gij(U), i
2.4. Estimation from time series data
FY→X|Z from9 a numerical time series
So far we have discussed time series data in terms of abstract
stochastic processes; that is, we have assumed that our empirical
time series data behaves like a realisation of some VAR process. We
now turn to the crucial issues of estimation, under this assump-
um.
tion, of
The first stage is to determine an appropriate model order for the
regression (1). This may be achieved via standard techniques such
as the Akaike or Bayesian information criteria, or cross-validation
(McQuarrie and Tsai, 1998). The idea of model order selection is to
balance the number of parameters (in the VAR case this is deter-
mined by the maximum lag p) so as to achieve the best model
fit—perhaps in a ML or error-minimisation sense—while at the same
time avoiding overfitting a finite data sequence.
u1, . . .,
=
u
The next stage is to obtain estimates of the model parame-
ters which maximise the likelihood function for the respective
VAR models (equivalently, minimise model error). As mentioned
(Section 1), the MVGC toolbox obviates the need to estimate the
reduced model parameters separately from the data. Here there is
a choice of techniques yielding estimates asymptotically equivalent
to the ML estimate, notably ordinary least squares (OLS) (Hamilton,
1994) and various multivariate extensions of Durbin recursion (fre-
quently under the banner of “LWR algorithms”) (Levinson, 1947;
Whittle, 1963; Wiggins and Robinson, 1965; Morf et al., 1978) –
see Appendix A.1, (A2). Once we have estimates of all relevant
parameters for both the full and reduced models, the G-causality
sample estimatorFY→X|Z(u) is obtained via (22) with the residuals
covariance matrices �xx, �
xx replaced by their respective estima-
tors ˆ�xx(u), ˆ�
xx(u). Another route to estimation of causalities in
the time domain is via spectral G-causality (Section 2.6).
An important but rarely considered issue in G-causality model
ut is a VAR (1), it
Xt will always be a well-defined
estimation is that, given that the full process
is not clear that the subprocess
8 Confusingly, the term “multivariate” G-causality is sometimes applied in the
literature to this case.
9 This section and the following applies equally to the unconditional case (Section
2.2); we need simply take
Z
to be empty; i.e. of dimension 0.
VAR. In fact condition (11) guarantees that it is.10 However, even
if the full VAR (1) is of finite order, the reduced VAR (16) will gen-
erally be of infinite order11 (similar remarks apply to the reduced
regression (21)). Since under the ML formalism the full and reduced
model orders should be the same, this implies that the appropriate
(finite) empirical model order p should really be estimated for the
reduced, rather than the full regression model. Failure to do so has
potentially serious implications for G-causal inference (Section 2.5
– see also Section 3.1.2). The MVGC toolbox overcomes this issue
in a natural way by only requiring estimation of the full regression
(Section 3.1).
2.5. Statistical inference
The next task is to establish the statistical significance of the
estimated causality against the null hypothesis (17) of zero causal-
ity, and, if desired, confidence intervals for its magnitude. As a
likelihood ratio test, standard large-sample theory (Wilks, 1938;
Wald, 1943) applies to the time-domain G-causality estimator in
X) = nx, dim(
both the conditional and unconditional cases. If dim(
Z) = nz (with nx + ny + nz = n) then the difference in
Y) = ny and dim(
the number of parameters between the full model (20) and the
≡
nested reduced model (21) is just d
pnxny. Thus under the null
−
hypothesis (17) of zero causality, (m
estimator scaled by sample size has an asymptotic 2(d) distri-
bution. Under the alternative hypothesis the scaled estimator has
an asymptotic noncentral-2(d ; ) distribution, with noncentrality
FY→X|Z equal to the scaled actual causality
parameter
(which may, for the purpose of constructing confidence intervals,
be replaced by its estimator).
p)FY→X|Z(u), the G-causality
=
(m
−
p)
In the case of a univariate causal target [i.e. dim(
esis, exp(FY→X|Z(u))
X) = nx = 1] an
alternative asymptotic sampling distribution is available for the
−
R2-like statistic exp(FY→X|Z)
1: namely, under the null hypoth-
−
1 scaled by d2/d1 has an asymptotic F(d1,
−
d2) distribution where d1 = pny and d2 = m
p(n + 1), and under the
alternative hypothesis an asymptotic noncentral-F(d1, d2 ; ) dis-
tribution where again is equal to the actual scaled value of the
causality. For small samples in particular, the F-distribution may
be preferable (it has a fatter tail than the corresponding 2 distri-
bution). The MVGC toolbox makes both tests available and defaults
to the F-test if the target variable is univariate.
Since G-causality is non-negative,FY→X|Z(u) will generally be
positively biased, and under the alternative hypothesis the non-
central asymptotic distributions will be more accurate the closer
the actual causality is to zero (Wald, 1943; Geweke, 1982). Some
techniques for dealing with bias are discussed in Section 3.4.
For very small samples the theoretical asymptotic distributions
may not be sufficiently accurate and nonparametric data-derived
empirical sampling distributions may be preferable for statistical
inference. For this purpose the MVGC toolbox supplies permutation
test (Anderson and Robinson, 2001) and non-parametric bootstrap
(Efron, 1982) routines for significance testing and computation of
confidence intervals respectively; the toolbox also features routi-
nes suitable for simulation of surrogate time series data [Appendix
A.1, (A3)].
Finally, if multiple causalities are to be simultaneously consid-
ered, in view of the multiple null hypotheses we should take into
account family-wise error rates or false discovery rates (Hochberg
10 As noted by Geweke (Geweke, 1982), since the full CPSD S() is assumed to
Xt alone, from which
satisfy condition (11), so too does the CPSD Sxx() of the process
Xt are square summable, and
it follows that the coefficients of the autoregression of
the reduced VAR is thus well-defined. See Geweke (1982, 1984) for full details.
11 In fact if
finite-order VARMA process.
U t is a finite-order VAR, then a subprocess
Xt will in general be a
�
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
55
and Tamhane, 1987); again, this functionality is available in the
MVGC toolbox.
2.6. G-causality in the frequency domain
A powerful feature of G-causality is that it may be decomposed
in a natural way by frequency (Geweke, 1982, 1984). The resulting
spectral G-causality integrates to the time-domain causality previ-
ously introduced, which may thus be considered an average over
all frequencies of the spectral causality. The decomposition in the
unconditional case is derived as follows: a split of
into sub-
processes
as in (12) induces a decomposition
X,
U
Y
=
S()
Sxx()
Sxy()
Syx() Syy()
(24)
of the cross-power spectral density (Section 2.1) and a similar
decomposition for the transfer function H(). Now Sxx() is just
the CPSD of the sub-process
∗ +
∗
X, and from (9) we may derive
2Re{Hxx()�xyHxy()
=
Hxx()�xxHxx()
+
Hxy()�yyHxy()
Sxx()
(25)
∗}
Following Geweke (1982), we note that in the special case that
�xy ≡
0, which may always be effected by a linear transformation
FY→X invariant (Barrett et al., 2010), (25) takes
of variables leaving
the simpler form
Sxx()
=
Hxx()�xxHxx()
∗ +
Hxy()�yyHxy()
∗
(26)
whereby the CPSD of
splits into an “intrinsic” term and a “causal”
term. This motivates definition of the (unconditional) spectral G-
causality from
to
as
X
Y
X
≡
ln
f Y→X()
or, in terms of untransformed variables (i.e. where �xy /=
|Sxx()
∗|
|Sxx()|
−
Hxy()�yyHxy()
0),
f Y→X()
≡
ln
|Sxx()|
−
Hxy()�y|xHxy()
|Sxx()
∗|
(27)
(28)
�yy −
where the partial covariance matrix �y|x is defined by
�y|x ≡
Geweke then establishes the fundamental spectral decomposition
of G-causality in the unconditional case12
−1
xx �xy
�yx�
(29)
f Y→X()d
=
FY→X
(30)
2
1
2
0
so that time domain causality may be thought of as the average
over all frequencies of spectral causality.
Sample estimation of
f Y→X() admits at least two possible
approaches. A straightforward procedure is to estimate parame-
ters Ak, � for the VAR(p) (1) as before (after selecting a suitable
model order). The transfer function may then by calculated by (fast)
Fourier transform of the estimated regression coefficients accord-
ing to (10), the CPSD from (9) and the partial covariance matrix
from (29). An estimate for the unconditional spectral G-causality
is then obtained by plugging the appropriate estimates into (28).
Note that no reduced regression is required. An alternative proce-
dure (Dhamala et al., 2008a,b) is to estimate the CPSD S() directly
fact equality
xx Axy()|
−1
−
12 In
≤.
�yx�
In practice, according to Geweke (1982), the equality condition is “almost always”
satisfied.
|Ayy()
≤
2; otherwise it should be replaced by
/=
0 is satisfied on 0 <
(30) holds strictly when
the condition
in
from the data – many standard techniques are available to this end.
Then, as mentioned previously (Section 2.1), H() and � may be
computed numerically and an estimate for the spectral G-causality
is again obtained from (28). The MVGC toolbox facilitates both
techniques; we recommend the former due to improved numerical
accuracy and computational efficiency (Section 3.1 and Appendix
A.1). As regards statistical inference, in contrast to the time-domain
case there are (to our knowledge) no known (asymptotic) distribu-
tions for the sample distribution of ˆf Y→X() (see Geweke, 1984 for
a fuller discussion on this issue) and nonparametric techniques are
best deployed for significance testing and derivation of confidence
intervals13.
The conditional case is less straightforward. Suppose that
U
Z
as per (19). We then consider the
X,
splits into sub-processes
reduced regression [cf. (21)]
Y,
Xt
Zt
p
k=1
=
to define the residuals
the identity
FY→X|Z ≡
FY⊕Z†→X†
xx,k A
A
xz,k
A
zx,k Azz,k
X†,
+
X†
Z†
t
t
Xt−k
Zt−k
(31)
(32)
(33)
(34)
Z† as new variables. In Geweke (1984)
t
Y
≡
⊕
Z†
is established, where
Y t
Z†
ditional time-domain G-causality
causality in terms of the new variables
the spectral causality in the unconditional case is defined as
f Y→X|Z()
, which expresses the con-
FY→X|Z as an unconditional
Z†. Accordingly,
f Y⊕Z†→X† ()
X†,
⊕
≡
Y
t
2
1
2
0
and the spectral decomposition
f Y→X|Z() d
=
FY→X|Z
again obtains.
in
separate
sample, a
To estimate ˆf Y→X|Z()
reduced
regression—which, as has been pointed out (Chen et al., 2006a),
may lead to substantial inaccuracies14—may be avoided as follows:
after estimation of VAR parameters for (1) (or spectral estimation),
it may be shown [Appendix A.1, (A11) and (A12)] that the trans-
Z† defined by (31) induces a
formation of variables
transformation of the autocovariance sequence and CPSD which
may be effected computationally. Then the conditional spectral
causality may be calculated as for the unconditional case via (33).
X|Z()
Again, little is known of the sampling distribution of
so that nonparametric techniques must be used for statistical
inference.
f Y →
X†,
→
X,
Z
As
in the time-domain case, pairwise-conditional spectral
causalities may be calculated as
gij(U; )
()
≡
→Ui
|U[ij]
f Uj
(35)
2.7. Filter invariance of G-causality
We have previously shown that multivariate G-causality, condi-
tional and unconditional and in both time and frequency domains,
causalityFY→X , although we do not yet have a theoretical justification for this claim.
13 Our unpublished simulation studies suggest that, in fact, the sampling distribu-
tion for ˆf Y→X () at any fixed frequency is actually the same as for the time-domain
This appears also to hold in the conditional case.
14 The “block decomposition” technique proposed in Chen et al. (2006a) appears,
according to our unpublished simulations, to yield inaccurate results. In particular,
the integral equality (34) generally fails even for simple test data in large samples.
It is possible that the analytic derivation in Chen et al. (2006a) may be flawed.
�
56
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
is theoretically invariant under the application of (almost) arbitrary
stable, invertible15 digital filters (Antoniou, 1993; Barnett and Seth,
2011). Empirically, however, filtering of stationary time series data
may severely compromise statistical inference of causalities esti-
mated in sample. This is due primarily to an increase in empirical
VAR model order following filtering. An important implication is
that G-causality restricted to particular frequency bands cannot be
measured by first filtering the data within the desired bands. While
the computed values may change following filtering, this likely
reflects not the desired “band-limited” G-causality but rather spuri-
ous values arising from inaccuracies in model fitting and statistical
inference. The solution proposed in Barnett and Seth (2011) is to
avoid filtering (of stationary data – see below) altogether. Then if
spectral causalities are required, values outside the frequency range
of interest may simply be ignored, while appropriate time-domain
causalities may be obtained by averaging spectral causalities (28)
B
and (33) over the frequency range
of prior interest [cf. (30) and
(34)] to obtain band-limited G-causality:
FY→X(B)
f Y→X() d
(36)
B
≡ 1
(B)
≡ 1
(B)
≡
FY→X|Z(B)
f Y→X|Z() d
B
(37)
B. Once again,
where (B)
nonparametric methods must be used for statistical inference of
band-limited G-causality.
B d is the measure (length) of
In practice, some filtering may still be appropriate in cases
where the data are not stationary to begin with, in which case G-
causal analysis is likely anyway to fail or deliver spurious results
(Section 3.3). In these cases filtering may be a valid and useful tool
to improve stationarity; e.g. notch-filtering of line-noise in elec-
tronically recorded time series data (Barnett and Seth, 2011), or
finite differencing to eliminate drift (Seth, 2010). In the latter case,
filters of the form ˜ut =
ut−1 may be applied iteratively until
(approximate) stationarity is achieved; but note that differencing
constitutes a non-invertible filter, so that stationary data should
not be differenced. An important application of filter invariance in
neuroscience (Section 4) is to G-causal analysis of fMRI (functional
Magnetic Resonance Imaging) data, where serious confounds have
been previously suspected; this is discussed further in Section 4.3.
ut −
3. MVGC Toolbox design
Central to the design of the MVGC toolbox is the equivalence
(Section 2.1) of the VAR parameters (Ak, �), the autocovari-
ance sequence �k and the cross-power spectral density S() as
representations for a VAR process. The MVGC toolbox exploits
these equivalences to provide numerically accurate algorithms
for moving flexibly between the alternative VAR representations,
thus furnishing computationally efficient pathways for calculation
of G-causalities, conditional and unconditional, in the time and
frequency domains. A schematic of computational pathways imple-
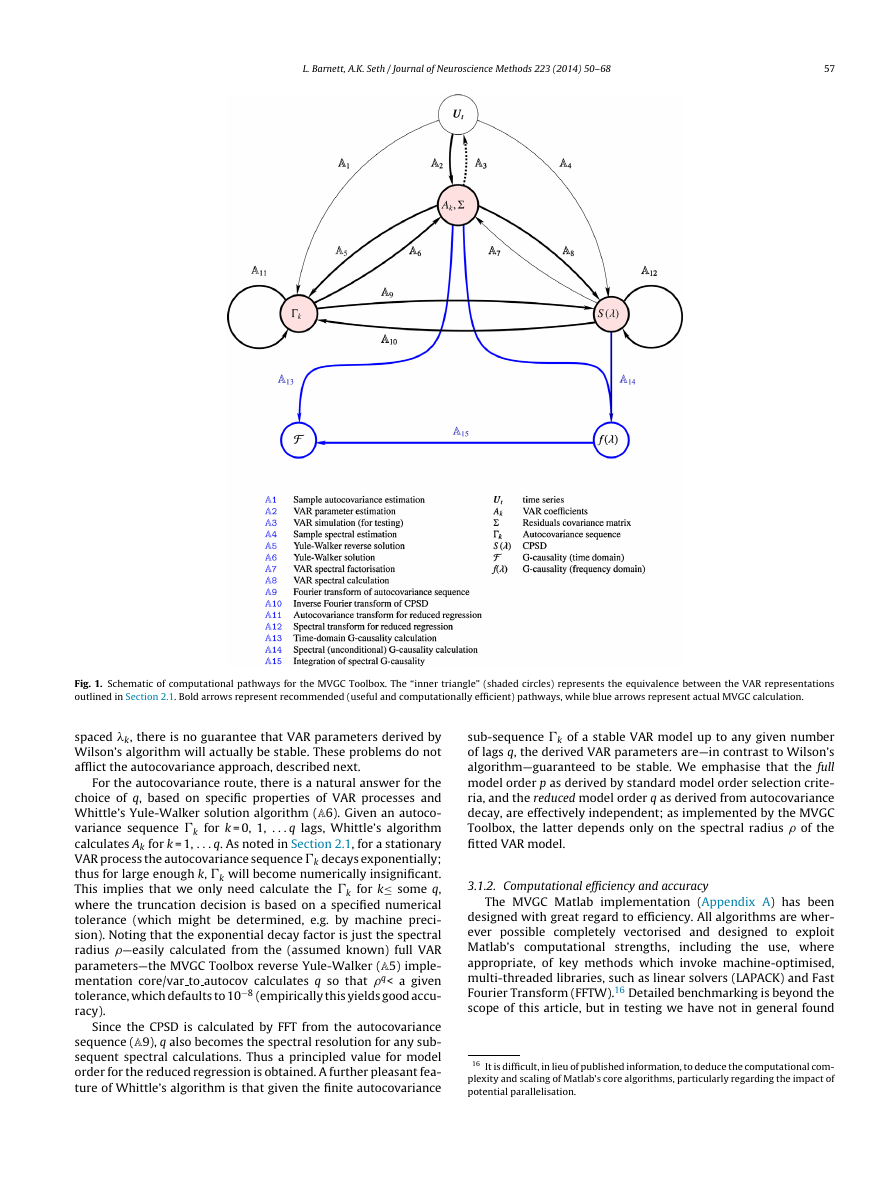
mented in the toolbox is given in Fig. 1.
3.1. Computational strategy
The basic operating principle of estimation of G-causalities via
the MVGC toolbox is that (after determination of a suitable model
15 In Barnett and Seth (2011) it is erroneously stated that to guarantee invariance
a filter need only be causal—i.e. not contain a pure lag. In fact full invertibility (min-
imum phase filtering) is required to ensure that the condition (11) is not violated
for the filtered process. We are grateful to Victor Solo (private communication) for
pointing this out.
order) a VAR (1) is fitted to the time series data just once (A2)
and all subsequent calculations are based on the estimated model
parameters ˆAk, ˆ�. This approach is theoretically consistent with
the fundamental assumption of G-causal estimation, that the time
series data is a realisation of a stationary VAR (1). In principle,
any of the equivalent VAR representations might be chosen for
initial estimation—e.g. via sample autocovariance (A1
A6) or
spectral estimation (A4
A7)—but empirically direct estimation
(A2) proves to be the most stable, accurate and computationally
efficient; nonetheless the MVGC toolbox allows implementation of
alternative methods.
→
→
Having acquired the VAR model, (conditional) G-causality in
the time (22) and frequency (33) domains involves estimation of
VAR parameters for the reduced regressions (21) and (31) respec-
tively. There is no simple algorithm (to our knowledge) to compute
reduced regression VAR parameters directly from full regression
parameters. Indeed, the standard approach [e.g. as implemented
in Seth (2010)] is to perform the reduced regressions explicitly
from the data (A2); however this leads to inaccuracies that may
be particularly serious in the spectral case (see Section 2.6 and
Chen et al., 2006a). Fortunately, numerical algorithms are available
for obtaining reduced regression parameters from the autocovari-
ance sequence (A7) via solution of the Yule-Walker equations (6),
or from the CPSD (A7) via spectral factorisation (9); the autoco-
variance sequence and CPSD may themselves be computed from
the (full) VAR parameters by “reverse” Yule-Walker solution (A5)
and straightforward spectral calculation (Fourier transform) (A8)
respectively. The recommended pathway for the MVGC Toolbox
is to use the autocovariance sequence. This is on the grounds
that, as revealed by empirical testing over a broad range of VAR
parameters, system size and model orders, Whittle’s algorithm
(A8) (Whittle, 1963) for Yule-Walker solution is faster, more stable
and more accurate than Wilson’s spectral factorisation algorithm
(A7) (Wilson, 1972); it also has additional useful properties (see
below). The toolbox thus uses the autocovariance sequence as the
dominant representation for all G-causality routines (A13,
A14)
although, again, the alternative spectral pathways may be imple-
mented if desired. Note that a useful “reality check” for the accuracy
of G-causality computations is that time-domain causalities should
integrate with reasonable accuracy to their spectral counterparts
(34) (A15).
3.1.1. Reduced model order, spectral resolution and
autocovariance lags
As noted previously (Section 2.4), even if the model order for
the full regression (20) is finite (and in practice a finite full model
order will, of course, need to be selected), the model order for the
reduced regressions (21) and (31) will be in theory infinite. If, as
described above, reduced VAR parameters are calculated from the
autocovariance sequence (or CPSD) how do we choose a suitable
finite model order?
First, we note that spectral factorisation (9) specifies the transfer
function H() at frequencies for which the CPSD S() is specified.
Now the VAR coefficients Ak are calculated by matrix inversion of
the transfer function followed by Fourier transform. Since the Fast
Fourier Transform (FFT) (Cooley and Tukey, 1965) will be used, this
implies that to calculate VAR coefficients from the CPSD, the CPSD
must be specified at some set of frequencies 1, . . . q evenly spaced
on [0, f], where f is the sampling frequency—indeed, Wilson’s algo-
rithm (A7) assumes this, and approximates the H(k) accordingly.
Then the Ak will naturally be derived for k = 1, . . ., q; i.e. the spec-
tral resolution q will, effectively, be the model order for the reduced
regression. Unfortunately, for the spectral factorisation approach
there is no obvious way to choose q. Furthermore (in contrast to
Whittle’s algorithm – see below), given a CPSD S() for a stable
VAR process evaluated, for a given spectral resolution q at evenly
�
L. Barnett, A.K. Seth / Journal of Neuroscience Methods 223 (2014) 50– 68
57
Fig. 1. Schematic of computational pathways for the MVGC Toolbox. The “inner triangle” (shaded circles) represents the equivalence between the VAR representations
outlined in Section 2.1. Bold arrows represent recommended (useful and computationally efficient) pathways, while blue arrows represent actual MVGC calculation.
spaced k, there is no guarantee that VAR parameters derived by
Wilson’s algorithm will actually be stable. These problems do not
afflict the autocovariance approach, described next.
For the autocovariance route, there is a natural answer for the
choice of q, based on specific properties of VAR processes and
Whittle’s Yule-Walker solution algorithm (A6). Given an autoco-
variance sequence �k for k = 0, 1, . . . q lags, Whittle’s algorithm
calculates Ak for k = 1, . . . q. As noted in Section 2.1, for a stationary
VAR process the autocovariance sequence �k decays exponentially;
thus for large enough k, �k will become numerically insignificant.
This implies that we only need calculate the �k for k≤
some q,
where the truncation decision is based on a specified numerical
tolerance (which might be determined, e.g. by machine preci-
sion). Noting that the exponential decay factor is just the spectral
radius —easily calculated from the (assumed known) full VAR
parameters—the MVGC Toolbox reverse Yule-Walker (A5) imple-
mentation core/var to autocov calculates q so that q< a given
tolerance, which defaults to 10−8 (empirically this yields good accu-
racy).
Since the CPSD is calculated by FFT from the autocovariance
sequence (A9), q also becomes the spectral resolution for any sub-
sequent spectral calculations. Thus a principled value for model
order for the reduced regression is obtained. A further pleasant fea-
ture of Whittle’s algorithm is that given the finite autocovariance
sub-sequence �k of a stable VAR model up to any given number
of lags q, the derived VAR parameters are—in contrast to Wilson’s
algorithm—guaranteed to be stable. We emphasise that the full
model order p as derived by standard model order selection crite-
ria, and the reduced model order q as derived from autocovariance
decay, are effectively independent; as implemented by the MVGC
Toolbox, the latter depends only on the spectral radius of the
fitted VAR model.
3.1.2. Computational efficiency and accuracy
The MVGC Matlab implementation (Appendix A) has been
designed with great regard to efficiency. All algorithms are wher-
ever possible completely vectorised and designed to exploit
Matlab’s computational strengths,
the use, where
appropriate, of key methods which invoke machine-optimised,
multi-threaded libraries, such as linear solvers (LAPACK) and Fast
Fourier Transform (FFTW).16 Detailed benchmarking is beyond the
scope of this article, but in testing we have not in general found
including
16 It is difficult, in lieu of published information, to deduce the computational com-
plexity and scaling of Matlab’s core algorithms, particularly regarding the impact of
potential parallelisation.
�















 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc