Brain tumor segmentation using deep learning
Gal Peretz , Elad Amar
Abstract
Brain tumor is one of the deadliest forms of cancer and as all cancer
types, early detection is very important and can save lives. By the time
symptoms appear cancer may have begun to spread and be harder to
treat. The screening process (the process of performing MRI or CT scans
to detect tumors) can be devided to two main tasks. The first task is the
classification task, doctors need to identify the type of the brain tumor.
There are three types of brain tumors menigioma, pituitray and glioma.
the type of the tumor can be an indication of the tumor‘s aggressiveness,
however to estimate the tumor’s stage (the size of the cancer and how
far it’s spread) an expert needs to segment the tumor first in order to
measure it in an accurate way. This lead to the second and more time
consuming task of segment the brain tumors which doctors need to seprate
the infected tissues from the healty ones by label each pixel in the scan.
This paper will investigate how can we utilize deep learning to help doctors
in the segmention process. The paper will be devided into four main
sections.
in the first section we will explore the problem domain and
the existing approachs of solving it. the second section will dicuss about
the UNet architecture and it‘s variations as this model gives state of the
art result on various medical image(MRI/CT) datasets for the semantic
segmentation task. In the third section we will describe how we chose to
adapt the Deep ResUnet architecture [7] and the experiments setup that
we did to evaluate our model.
In addition we will introduce the ONet
architecture and show how we can boost the model performance by using
bounding box labels.
1
Introduction
Cancer is one of the leading causes of death globally and it is responsible for 9.6
million deaths a year. One of the most deadliest type of cancer is brain cancer,
the 5-years survival rate is 34% for men and 36% for women. An prevalent
treatment for brain tumors is radiation threapy where high-energy radiation
source like gamma rays or x-rays shoots in a very precise way at the tumor and
therefore kill the tumor‘s cells while sparing surrounding tissues. However in
order to perfom the radiation treatment doctors need to segment the infected
tissues by separate the infected cells from the healthy ones. Creating this
segmentation map in an accurate way is very tedious, expensive, time-consuming
and error-prone task therefore we can gain a lot from automate this process.
1
�
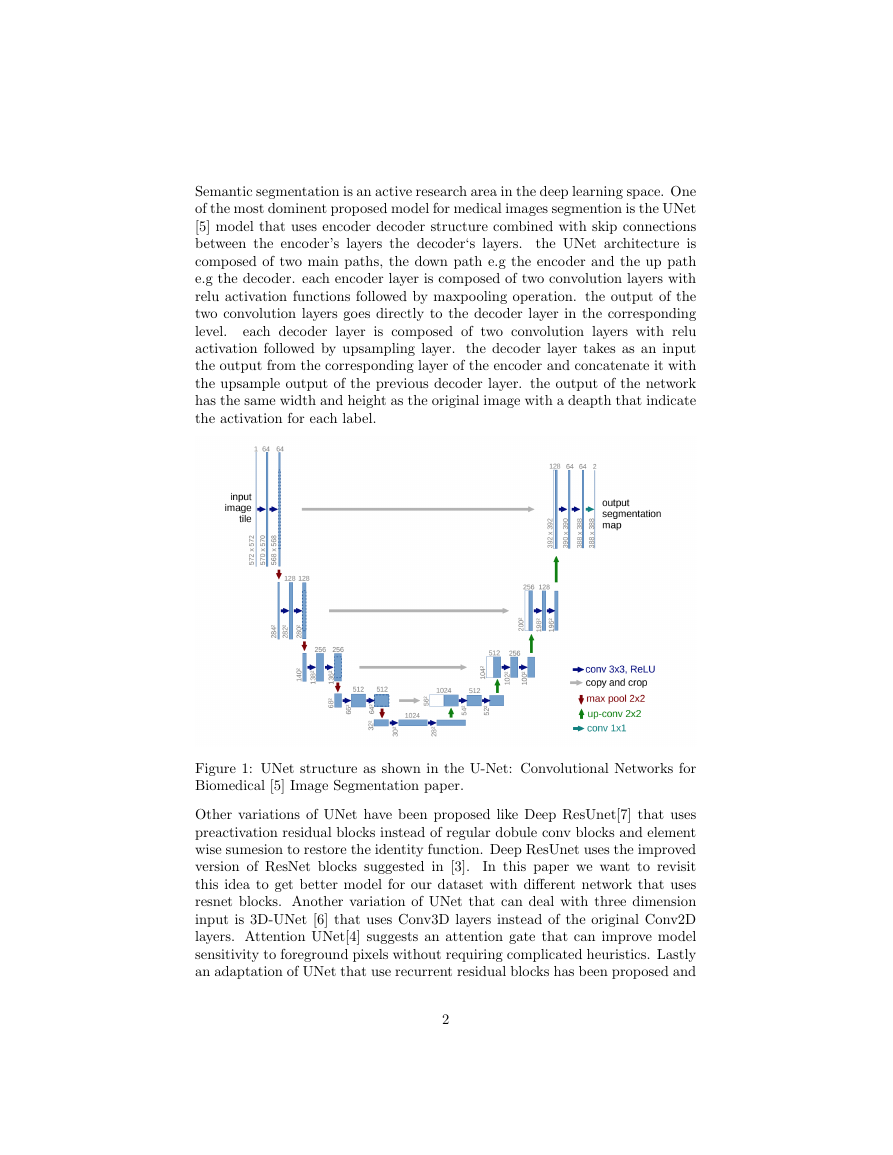
Semantic segmentation is an active research area in the deep learning space. One
of the most dominent proposed model for medical images segmention is the UNet
[5] model that uses encoder decoder structure combined with skip connections
between the encoder’s layers the decoder‘s layers.
the UNet architecture is
composed of two main paths, the down path e.g the encoder and the up path
e.g the decoder. each encoder layer is composed of two convolution layers with
relu activation functions followed by maxpooling operation. the output of the
two convolution layers goes directly to the decoder layer in the corresponding
level.
each decoder layer is composed of two convolution layers with relu
activation followed by upsampling layer. the decoder layer takes as an input
the output from the corresponding layer of the encoder and concatenate it with
the upsample output of the previous decoder layer. the output of the network
has the same width and height as the original image with a deapth that indicate
the activation for each label.
Figure 1: UNet structure as shown in the U-Net: Convolutional Networks for
Biomedical [5] Image Segmentation paper.
Other variations of UNet have been proposed like Deep ResUnet[7] that uses
preactivation residual blocks instead of regular dobule conv blocks and element
wise sumesion to restore the identity function. Deep ResUnet uses the improved
version of ResNet blocks suggested in [3].
In this paper we want to revisit
this idea to get better model for our dataset with different network that uses
resnet blocks. Another variation of UNet that can deal with three dimension
input is 3D-UNet [6] that uses Conv3D layers instead of the original Conv2D
layers. Attention UNet[4] suggests an attention gate that can improve model
sensitivity to foreground pixels without requiring complicated heuristics. Lastly
an adaptation of UNet that use recurrent residual blocks has been proposed and
2
�
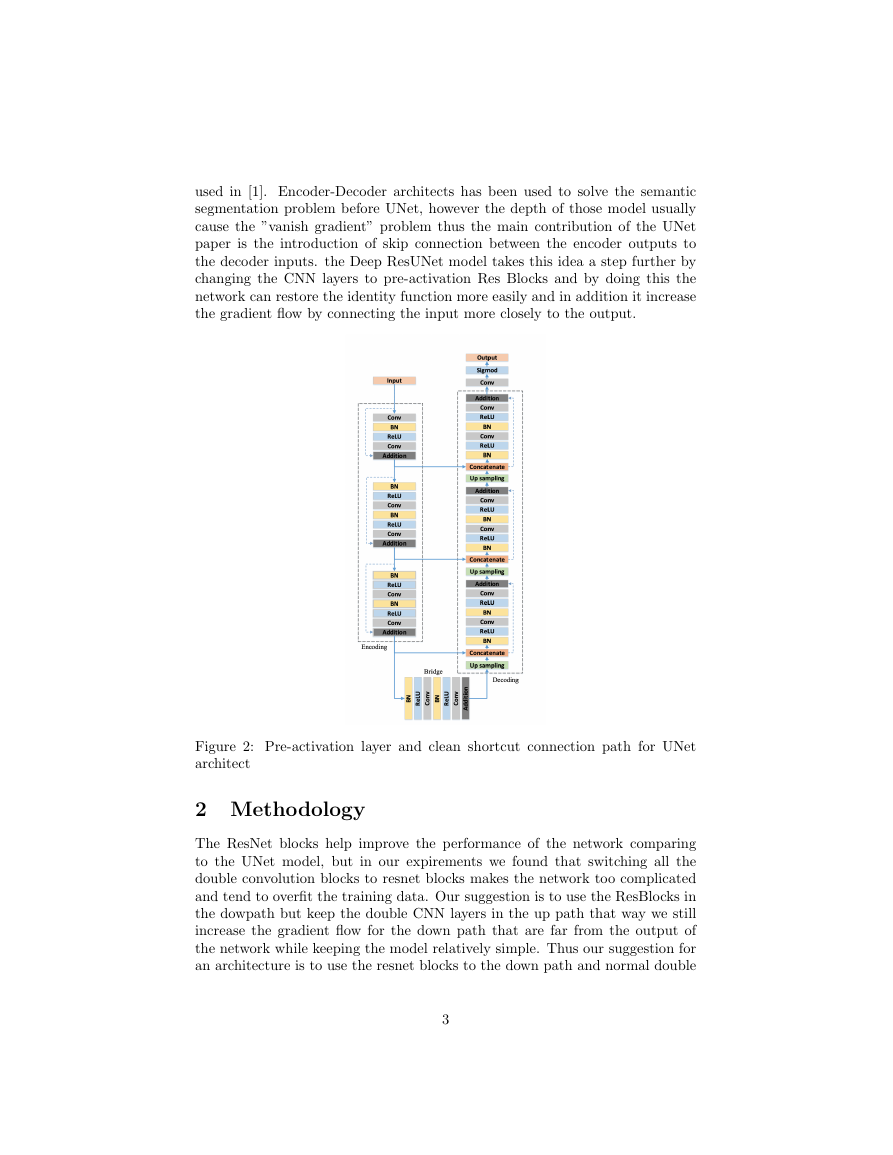
used in [1]. Encoder-Decoder architects has been used to solve the semantic
segmentation problem before UNet, however the depth of those model usually
cause the ”vanish gradient” problem thus the main contribution of the UNet
paper is the introduction of skip connection between the encoder outputs to
the decoder inputs. the Deep ResUNet model takes this idea a step further by
changing the CNN layers to pre-activation Res Blocks and by doing this the
network can restore the identity function more easily and in addition it increase
the gradient flow by connecting the input more closely to the output.
Figure 2: Pre-activation layer and clean shortcut connection path for UNet
architect
2 Methodology
The ResNet blocks help improve the performance of the network comparing
to the UNet model, but in our expirements we found that switching all the
double convolution blocks to resnet blocks makes the network too complicated
and tend to overfit the training data. Our suggestion is to use the ResBlocks in
the dowpath but keep the double CNN layers in the up path that way we still
increase the gradient flow for the down path that are far from the output of
the network while keeping the model relatively simple. Thus our suggestion for
an architecture is to use the resnet blocks to the down path and normal double
3
�
convolution block to the up path we will call this model the hybrid ResUnet. we
will show that this model generalize better and get higher performance on the
dice matric for our dataset. we use the ”brain tumor dataset” [2], this dataset
consist of 3064 MRI scans represented as 512 x 512 matrics, and 512 x 512
boolean masks that indicate the pixels of the infected tissues in the image. Our
performace metric will be the dice coefficient as this is a common metric for
the segmentation problem, when there is imbalance between the true labels(the
tumor‘s pixels) and the false labels(the non tumor‘s pixels). the dice formula
for the binary case can be stated as follows:
2T P
2T P + F P + F N
And more concrete in our case the network will output 512 x 512 score map
then after softmax and thresholding we will convert it to 0-1 map. we will check
the set similarity of this map with the corresponding labeled mask using the
dice formula as such:
Let X := output map
Let Y := labeled mask
2 · |X ∩ Y |
|X| + |Y |
For the loss function we will use the dice loss, the dice loss is a soft version of
the dice formula and gives a value between 0 to 1, where 0 means perfect match
between the sets.
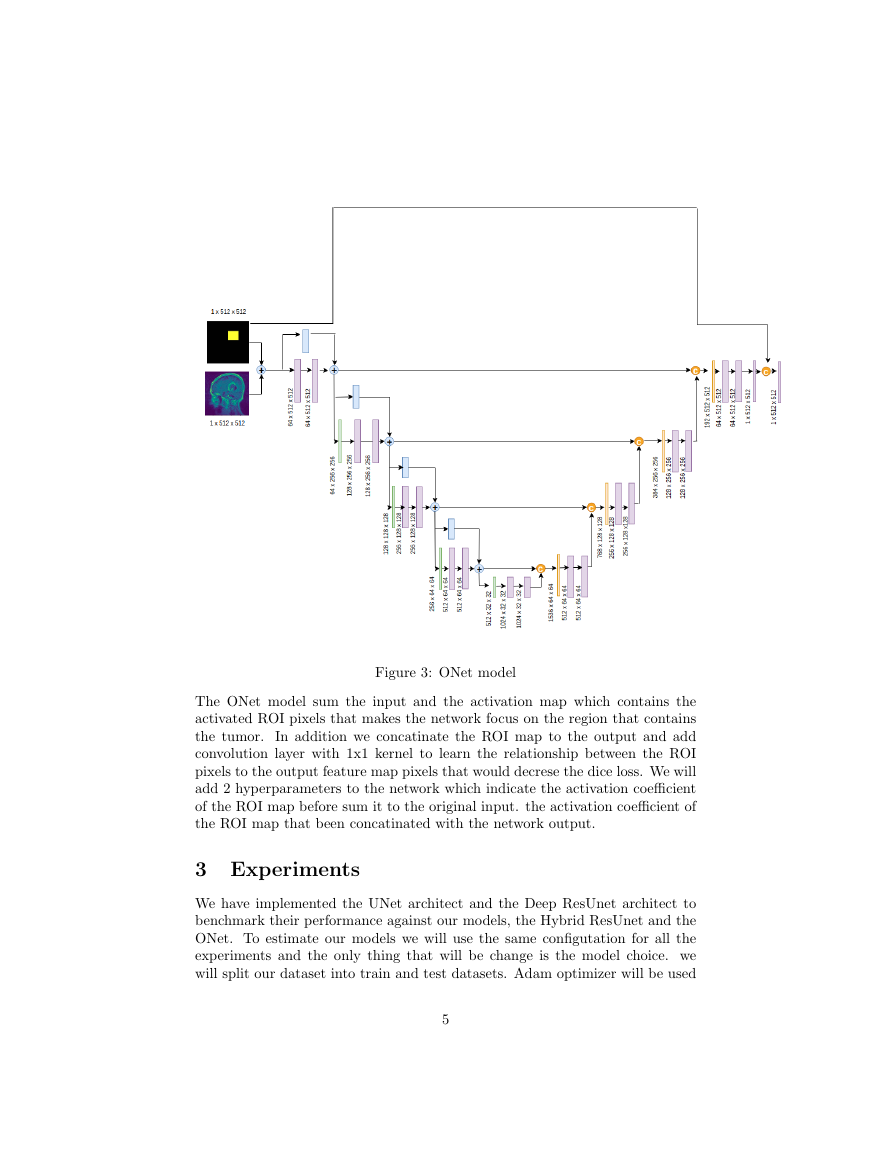
We can think of a way to provide additional knowledge to perform the semantic
task better by adding ”regions of interest (ROI)” to the network input and
”inject” it to the network in the right places. Classic computer vision algorithms
can be use to estimate the ROI or we can ask experts to provide rough estimation
of the tumor region by drawing a bounding box around it(or give us 4 dimensional
vector [x min, x max, y min, y max]). The suggested network use skip connection
with concatination that makes the UNet architecht looks like ONet so we will
use this name as an alias to the proposed model.
4
�
Figure 3: ONet model
The ONet model sum the input and the activation map which contains the
activated ROI pixels that makes the network focus on the region that contains
the tumor.
In addition we concatinate the ROI map to the output and add
convolution layer with 1x1 kernel to learn the relationship between the ROI
pixels to the output feature map pixels that would decrese the dice loss. We will
add 2 hyperparameters to the network which indicate the activation coefficient
of the ROI map before sum it to the original input. the activation coefficient of
the ROI map that been concatinated with the network output.
3 Experiments
We have implemented the UNet architect and the Deep ResUnet architect to
benchmark their performance against our models, the Hybrid ResUnet and the
ONet. To estimate our models we will use the same configutation for all the
experiments and the only thing that will be change is the model choice. we
will split our dataset into train and test datasets. Adam optimizer will be used
5
�
with learning rate of 0.001 for all of the experiments and the loss function will
be a soft version of the dice metric as known as dice loss. The input will be
512 x 512 grayscale images (one channel) and the batch size will be 2 images.
we want to retain the high resolution of the images because this is essential
for the segmentation task. we normalized the images by subtracting the mean
of the images. The first experiment will compare the dice performance of the
UNet, Deep ResUnet and the Hybrid solution where the down path contains
Res blocks and the up path double conv blocks. The second experiment will
compare the ONet to the other networks.
4 Result
First we will analyze the Hybrid ResUnet model generalization capability by
comparing it with the UNet architecture and the ResUnet architecture with the
same experiment setup as described above.
Figure 4: First experiment dice score results
Model
UNet
Deep ResUnet
Hybrid ResUnet
Dice score
0.8098
0.8318
0.8366
we can notice that the hybrid solution between resnet blocks and double
conv blocks genralization better than the basic Unet and the Deep ResUnet for
our dataset and in addition it convert faster and has less noisy curve.
6
�
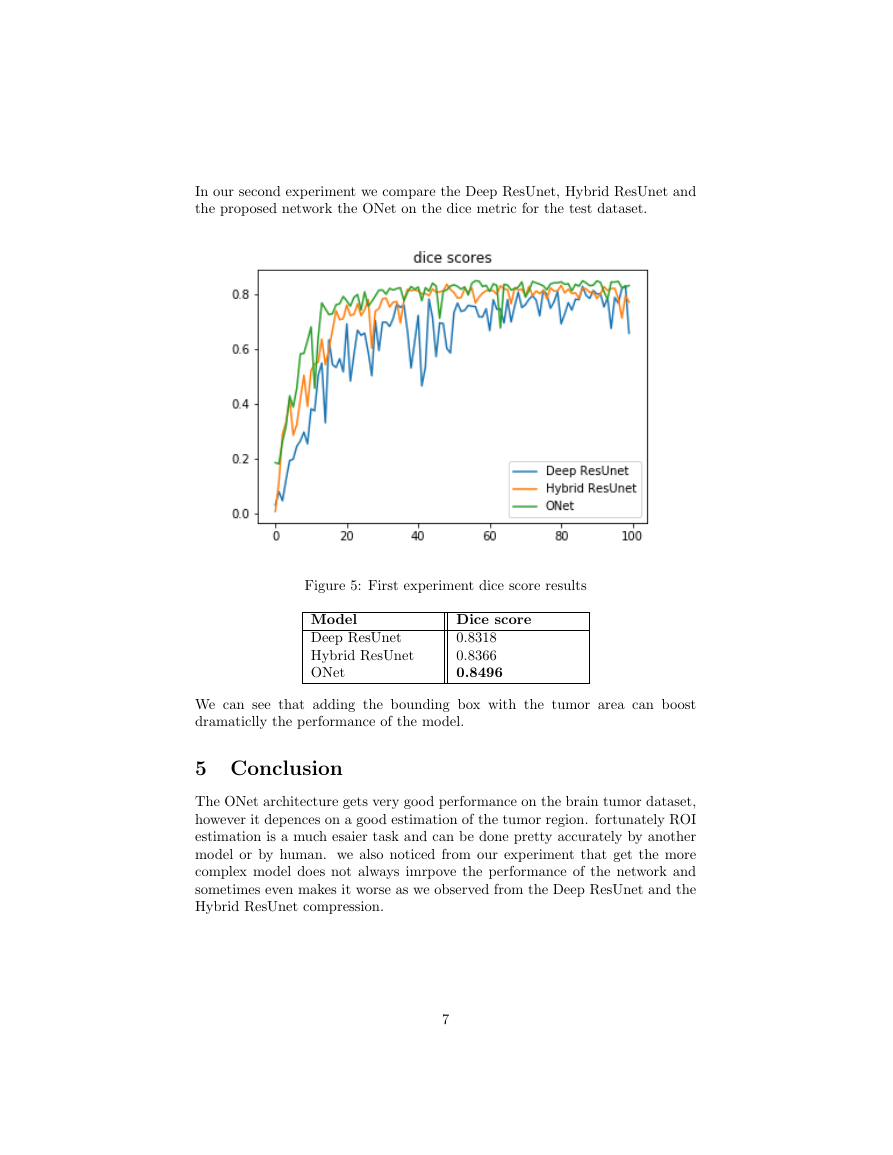
In our second experiment we compare the Deep ResUnet, Hybrid ResUnet and
the proposed network the ONet on the dice metric for the test dataset.
Figure 5: First experiment dice score results
Model
Deep ResUnet
Hybrid ResUnet
ONet
Dice score
0.8318
0.8366
0.8496
We can see that adding the bounding box with the tumor area can boost
dramaticlly the performance of the model.
5 Conclusion
The ONet architecture gets very good performance on the brain tumor dataset,
however it depences on a good estimation of the tumor region. fortunately ROI
estimation is a much esaier task and can be done pretty accurately by another
model or by human. we also noticed from our experiment that get the more
complex model does not always imrpove the performance of the network and
sometimes even makes it worse as we observed from the Deep ResUnet and the
Hybrid ResUnet compression.
7
�
References
[1] Md Zahangir Alom et al. “Recurrent Residual Convolutional Neural Network
based on U-Net (R2U-Net) for Medical Image Segmentation”. In: arXiv
(Feb. 2018). eprint: 1802.06955. url: https://arxiv.org/abs/1802.
06955.
brain tumor dataset. [Online; accessed 27. Sep. 2019]. Apr. 2017. url:
https://figshare.com/articles/brain_tumor_dataset/1512427/5.
[2]
[3] Kaiming He et al. “Identity Mappings in Deep Residual Networks”. In:
arXiv (Mar. 2016). eprint: 1603.05027. url: https://arxiv.org/abs/
1603.05027.
[4] Ozan Oktay et al. “Attention U-Net: Learning Where to Look for the
Pancreas”. In: arXiv (Apr. 2018). eprint: 1804 . 03999. url: https : / /
arxiv.org/abs/1804.03999.
[5] Olaf Ronneberger, Philipp Fischer, and Thomas Brox. “U-Net: Convolutional
Networks for Biomedical Image Segmentation”. In: arXiv (May 2015).
eprint: 1505.04597. url: https://arxiv.org/abs/1505.04597.
[6] Chengjia Wang et al. “A two-stage 3D Unet framework for multi-class
segmentation on full resolution image”. In: arXiv (Apr. 2018). eprint: 1804.
04341. url: https://arxiv.org/abs/1804.04341.
[7] Zhengxin Zhang, Qingjie Liu, and Yunhong Wang. “Road Extraction by
Deep Residual U-Net”. In: arXiv (Nov. 2017). doi: 10.1109/LGRS.2018.
2802944. eprint: 1711.10684.
8
�












 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc