Pooling in Graph Convolutional Neural Networks
Mark Cheung
John Shi
Lavender Jiang
Electrical and Computer Engineering
Electrical and Computer Engineering
Electrical and Computer Engineering
Carnegie Mellon University
Pittsburgh, PA, USA
markcheu@andrew.cmu.edu
Carnegie Mellon University
Pittsburgh, PA, USA
jshi3@andrew.cmu.edu
Carnegie Mellon University
Pittsburgh, PA, USA
yaoj@andrew.cmu.edu
Oren Wright
Software Engineering Institute
Carnegie Mellon University
Pittsburgh, PA, USA
owright@sei.cmu.edu
Jos´e M.F. Moura
Electrical and Computer Engineering
Carnegie Mellon University
Pittsburgh, PA, USA
moura@andrew.cmu.edu
0
2
0
2
r
p
A
7
]
P
S
.
s
s
e
e
[
1
v
9
1
5
3
0
.
4
0
0
2
:
v
i
X
r
a
Abstract—Graph convolutional neural networks (GCNNs) are
a powerful extension of deep learning techniques to graph-
structured data problems. We empirically evaluate several pool-
ing methods for GCNNs, and combinations of those graph pooling
methods with three different architectures: GCN, TAGCN, and
GraphSAGE. We confirm that graph pooling, especially DiffPool,
improves classification accuracy on popular graph classification
datasets and find that, on average, TAGCN achieves comparable
or better accuracy than GCN and GraphSAGE, particularly for
datasets with larger and sparser graph structures.
Index Terms—graph convolutional neural network, graph
pooling, TAGCN, graph classification, graph signal processing
I. INTRODUCTION
Over the past decade, deep learning techniques such as
convolutional neural networks (CNNs) have transformed fields
like computer vision and other Euclidean data domains (i.e.,
domains in which data have a uniform, grid-like structure).
Many important domains, however, are comprised of non-
Euclidean data (i.e., data have irregular relationships that
require mathematical concepts like graphs or manifolds to ex-
plicitly model). Such domains include social networks, sensor
feeds, web traffic, supply chains, and biological systems. As
these data grow in size and complexity, deep learning seems
to recommend itself as a tool for classification and pattern
recognition, but conventional deep learning approaches are
often sharply limited when data lack a Euclidean structure to
exploit. There are ongoing efforts to extend deep learning to
these non-Euclidean domains, and such techniques have been
dubbed geometric deep learning [1].
In parallel with advances in geometric deep learning are
advances in graph signal processing (GSP) [2], [3]. Research
in GSP attempts to generalize classical signal processing
theory for irregular data defined on graphs. One attraction
This material is based upon work partially funded and supported by the
Department of Defense under Contract No. FA8702-15-D-0002 with Carnegie
Mellon University for the operation of the Software Engineering Institute, a
federally funded research and development center. This work is also partially
supported by NSF grants CCF 1837607 and CCN 1513936.
of GSP is that it provides a unified mathematical framework
through which to view the spectral and vertex domains of a
graph. Concepts like frequency or smoothness, which can be
understood intuitively in classical signal processing, can be
explicitly defined for data on graphs.
Graph convolutional neural networks (GCNNs), an exten-
sion of CNNs to graph-structured data, were first implemented
with concepts from spectral graph theory [4], and methods
based on the spectral approach have since been refined and
expanded [5], [6]. Reference [7] proposes the topology adap-
tive graph convolutional network (TAGCN) that defines graph
convolution directly in the vertex domain as multiplication by
polynomials of the graph adjacency matrix. This is consistent
with the concept of convolution in graph signal processing
[2]. TAGCN designs a set of fixed-size learnable filters whose
topologies are adaptive to the topology of the graph as the
filters scan the graph to perform convolution, see also [8], [9].
Other implementations, such as GraphSAGE [10] and graph
attention networks (GATs) [11], are also defined directly in the
vertex domain of the graph and apply a learned, convolution-
like aggregation function.
An important operation in conventional CNNs is pooling, a
nonlinear downsampling operation. Pooling layers in a CNN
shrink the number of dimensions of the feature representation,
thereby reducing the computation cost, memory footprint, and
number of learned parameters. As a result, pooling allows for
deeper networks in practice and can help control overfitting.
Additionally, pooling has translation invariance properties that
are desirable in many applications. Recently, the use of pooling
in CNNs has come into question, but it remains popular.
Just as convolution and convolution-like methods have been
proposed to create graph convolutional layers in GCNNs, sev-
eral methods have been proposed in order to perform pooling
with GCNNs [12], [13], [14]. Unlike convolution, which has
been derived in GSP [2], pooling has not been rigorously
defined. Therefore, the current generation of pooling methods
are based on ad hoc rather than systematic approaches. They
nonetheless have shown improved accuracy on popular graph
�
classification datasets.
In this paper, we perform experiments on graph classifica-
tion datasets, conditionally on graph convolution and graph
pooling in GCNNs. This is a supervised learning task in
which previously unseen graphs are classified based on labeled
graphs. This task is analogous to image classification. Like
with CNNs and image classification, tools like pooling layers
are important for constructing high-level representations from
node-level information.
The paper is divided as follows: we first present the back-
ground and related work in section II. Section III provides our
proposed approach. In section IV, we discuss the datasets used
and present the results and analysis. Finally, we conclude the
paper in section V.
A. Graph Signal Processing Perspective
The convolutional and pooling operator in graph neural
network have a theoretical foundation in GSP. GSP [2] extends
traditional discrete signal processing to graph signals, signals
that are indexed by the nodes in a graph.
Let G(A) = (V, E) be a graph with adjacency matrix A ∈
CN×N , where V is the set of N nodes and a nonzero entry
[A]ij denotes a directed edge eij ∈ E from node i to node j.
s : V → S on G is a graph signal where S is the signal space
over the nodes of G and S ⊆ CN . s = (s1, s2, ..., sN ) ∈ CN ,
and si represents a measurement at node vi ∈ V.
The heart of GCNNs is applying convolutional filters to
graph signals. In GSP, convolution is a matrix-vector multi-
plication of a polynomial of the adjacency matrix P (A) and
the graph signal x. This definition is used to create the graph
convolutional layer in GCNNs.
The GSP literature includes [15] and [16]. In [15] and
[16], several sampling set selection and sampling methods
are proposed. The pooling methods explored herein are not
based specifically on these sampling methods, but we observe
that there is a relationship between sampling in GSP and
pooling in GCNNs. Both reduce the number of values in
the signal and can reduce the number of nodes in the graph.
The key difference is that, in sampling, we focus on how to
recover the original signal given the sampled signal. However,
recoverability is not required in pooling algorithms in GCNNs.
II. RELATED WORK
In this section, we describe the infrastructure for graph
convolutional and pooling layers and the related literature.
A. Graph Convolutional Layer
We concentrate on three implementations of GCNNs, de-
rived from different definitions of graph convolution: graph
convolutional networks (GCNs) [6], GraphSAGE [10], and
topology-adaptive graph convolutional networks (TAGCNs)
[7], [9].
In GCN [6], given a graph signal X (0) ∈ Rn×c (where 0
denotes the input layer, n is the number of nodes, and c is
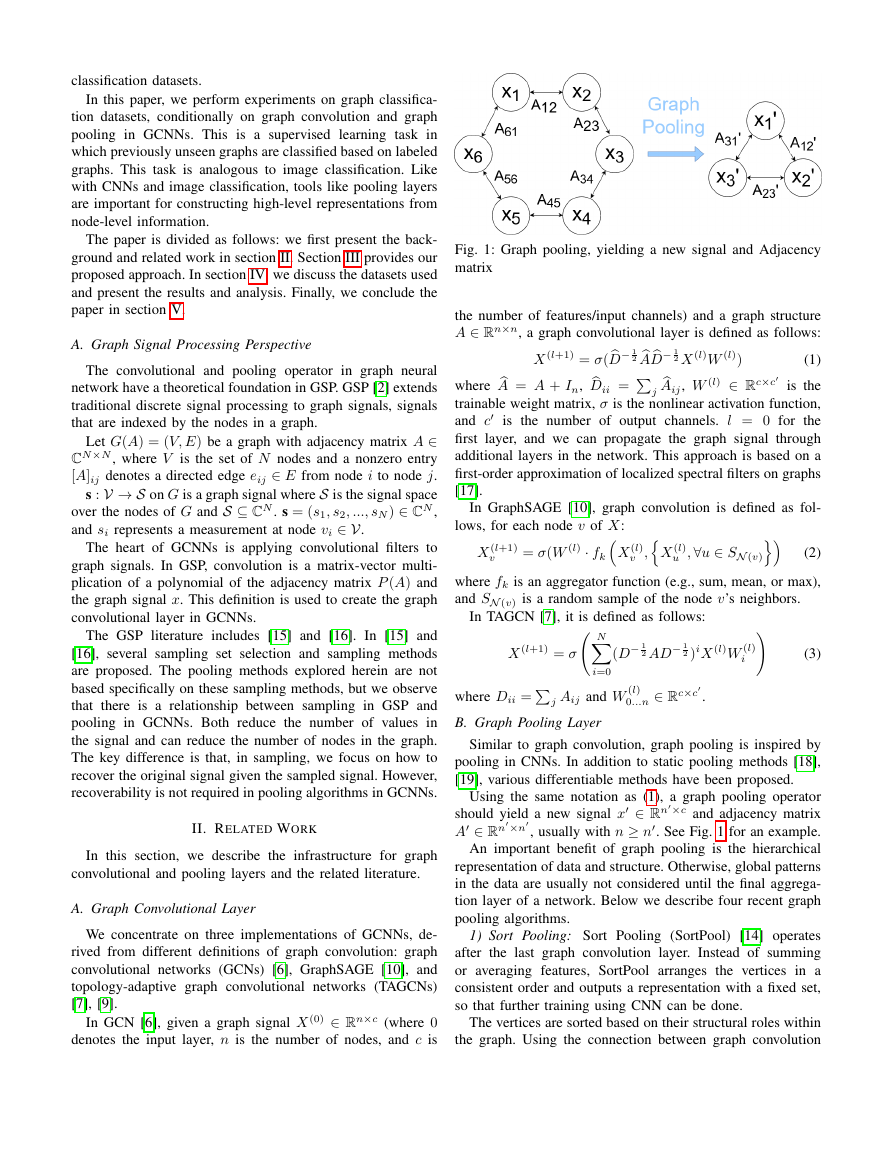
Fig. 1: Graph pooling, yielding a new signal and Adjacency
matrix
2 X (l)W (l))
the number of features/input channels) and a graph structure
A ∈ Rn×n, a graph convolutional layer is defined as follows:
(1)
2 AD− 1
X (l+1) = σ(D− 1
where A = A + In, Dii =
j Aij, W (l) ∈ Rc×c
is the
trainable weight matrix, σ is the nonlinear activation function,
and c
is the number of output channels. l = 0 for the
first layer, and we can propagate the graph signal through
additional layers in the network. This approach is based on a
first-order approximation of localized spectral filters on graphs
[17].
In GraphSAGE [10], graph convolution is defined as fol-
lows, for each node v of X:
X (l+1)
v
= σ(W (l) · fk
X (l)
v ,
where fk is an aggregator function (e.g., sum, mean, or max),
and SN (v) is a random sample of the node v’s neighbors.
In TAGCN [7], it is defined as follows:
u ,∀u ∈ SN (v)
X (l)
(2)
(3)
N
i=0
X (l+1) = σ
where Dii =
j Aij and W (l)
0...n ∈ Rc×c
.
(D− 1
2 AD− 1
2 )iX (l)W (l)
i
B. Graph Pooling Layer
Similar to graph convolution, graph pooling is inspired by
pooling in CNNs. In addition to static pooling methods [18],
[19], various differentiable methods have been proposed.
Using the same notation as (1), a graph pooling operator
should yield a new signal x ∈ Rn×c and adjacency matrix
A ∈ Rn×n
, usually with n ≥ n. See Fig. 1 for an example.
An important benefit of graph pooling is the hierarchical
representation of data and structure. Otherwise, global patterns
in the data are usually not considered until the final aggrega-
tion layer of a network. Below we describe four recent graph
pooling algorithms.
1) Sort Pooling: Sort Pooling (SortPool) [14] operates
after the last graph convolution layer. Instead of summing
or averaging features, SortPool arranges the vertices in a
consistent order and outputs a representation with a fixed set,
so that further training using CNN can be done.
The vertices are sorted based on their structural roles within
the graph. Using the connection between graph convolution
�
and the Weisfeiler-Lehman subtree kernel [20], SortPool sorts
the node features of the last layer individually, then sorts in
descending order based on the layer before, and finally selects
the top k nodes.
2) Differentiable Pooling: Differentiable Pooling (Diff-
Pool) [12] is a differentiable graph pooling module that learns
hierarchical representations of the graphs by aggregating nodes
through several pooling layers. It uses a learned assignment
matrix S ∈ Rn1×n2 and updates the graph signal and topology
as follows:
Z (l)
A(l)S(l)
X (l+1) = S(l)T
A(l+1) = S(l)T
(4)
(5)
where Z (l) ∈ Rn1×n1 is the GraphSAGE [10] operator with
the mean aggregator. DiffPool achieves significantly better
prediction accuracy than GraphSAGE, SortPool, and certain
kernel methods, especially when global features are important
for classification [12].
3) Top-k Pooling: Top-k Pool [13] pools using a trainable
projection vector p(l) ∈ Rf and select the top-k indices of the
projection X (l)p(l) and the corresponding edges in A(l).
Top-k pool is inspired by encoder-decoder architectures like
U-Nets. In addition to the Top-k pool operation, there is also
an Unpool operation that reverses the process. These two
combined create the encoder-decoder model on graph, known
as the graph U-Nets [13]. Reference [13] shows that Top-k
pool with the U-net structure performs better than DiffPool,
but we will show if it works well standalone vs. other pooling
algorithms.
4) Self-Attention Graph Pooling: Self-Attention Graph
Pooling (SagPool) [13] uses an attention mechanism to select
the important nodes:
y = GNN(X (l), A(l))
i = topk(y)
X (l+1) = (X (l) tanh(y))i
A(l+1) = A(l)
i,i
(6)
(7)
(8)
(9)
The attention score is calculated from GCN and the top
k nodes are selected from it. Since graph convolution is
used to obtain the self-attention score, SagPool uses both the
graph features and structure [13]. Reference [13] shows that
SAGPool performs better than DiffPool and Top-k Pool across
some biochemical datasets.
III. PROPOSED METHOD
We first compare GCN, GraphSAGE, and TAGCN for
graph classification across four benchmark datasets. We then
investigate how pooling affects these results, by combining
the different convolutional architectures with the four pooling
techniques described above, i.e., SortPool [14], DiffPool [12],
Top-k Pool [13], and SagPool [21]. In each instance, the pool-
ing method is paired with GCN or GraphSAGE (determined
by that used in each pooling paper), and compared with the
pooling method paired with TAGCN.
IV. EXPERIMENTS
A. Datasets
To evaluate the efficacy of the different methods, we apply
our methods on real-world graph kernel benchmarks. See Ta-
ble I for the properties of these datasets. We evaluate our meth-
ods on bioinfomatics datasets and social network datassets.
Both MUTAG and Proteins datasets are bioinformatics data.
MUTAG [22] is a dataset consisting of chemical compounds
represented by graphs. The task is to predict whether the
chemical compound is mutagenic. Proteins [23] is a dataset
consisting of proteins represented by graphs. The objective is
to predict whether a protein functions as an enzyme. In both of
the datasets, the nodes are structure elements, and two nodes
are connected if there is a chemical bond between the structure
elements represented by the nodes.
For social network datasets, we chose IMDB-Binary and
Reddit-Binary. IMDB-Binary [24] is a set of graphs corre-
sponding to ego-networks of actors and actresses. An edge is
drawn between two actors if they were cast in the same movie.
The task is to predict whether a movie is romance or action.
In Reddit-Binary [24], each graph corresponds to an online
discussion thread. An edge is drawn between two users if one
has replied to the other. The task is to predict whether a thread
belongs to a discussion forum or a question answering forum.
TABLE I: Properties of Graph Classification Datasets
Dataset
MUTAG
Proteins
IMDB-Binary
Reddit-Binary
Graphs
188
1113
1000
2000
Classes
2
2
2
2
Avg Nodes
17.7
39.06
19.77
429.63
Avg Edges
38.9
72.82
96.53
497.75
B. Network Training
We perform 5-fold cross-validation to select
the hyper-
parameters from the validation accuracy and estimate the
test accuracy. For the baselines, the hyperparameters are the
number of graph convolutional layers, number of channels in
each layer, dropout rates, pooling rate (number or percentage
of nodes to keep), and (for TAGCN) order of polynomial filter.
For a fairer comparison, we considered 1-5 layers for TAGCN
vs. 1-15 layers for GCN and GraphSAGE when using graph
polynomial filters of degree 3 (to show that 1 layer of TAGCN
with degree n is not n layers of GCN/GraphSAGE). We use
cross-entropy loss and ADAM optimization with a starting
learning rate of 0.01, a decay factor of 0.5, and a decay step
size of 50. Experiments were performed in PyTorch using code
from the PyTorch Geometric Library [25].
C. Results
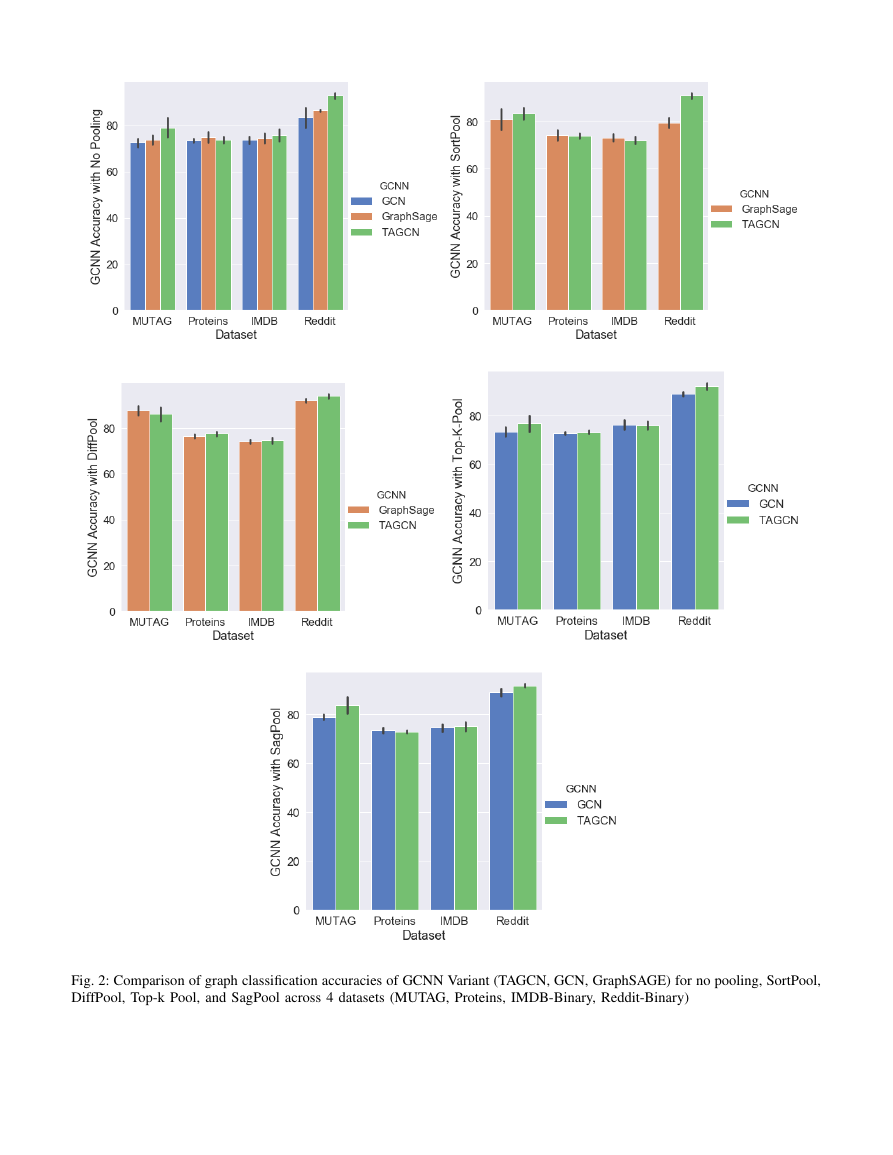
Fig. 2 shows the results of GCNN variants with no pooling,
DiffPool, SagPool, SortPool, and Top-K Pool. The green,
orange, and blue bars are the means of the cross-validated
accuracy and the smaller black error bars are their standard
deviations.
�
Fig. 2: Comparison of graph classification accuracies of GCNN Variant (TAGCN, GCN, GraphSAGE) for no pooling, SortPool,
DiffPool, Top-k Pool, and SagPool across 4 datasets (MUTAG, Proteins, IMDB-Binary, Reddit-Binary)
�
1) Graph Convolution Comparison:
In general, TAGCN
performs better than GCN and GraphSAGE on the four graph
classification benchmarks. However, due to the increase in
complexity, TAGCN has high variance, especially denser graph
structures. TAGCN performs better as graphs become less
sparse, i.e., as average degree increases.
We also showed empirically that simply increasing num-
ber of layers in GCN and GraphSAGE is not analogous to
increasing the order of the polynomial filter in TAGCN. We
attribute the the advantage of TAGCN mainly to: 1) Passing a
residual connection of the graph signal, and 2) Having weights
associated with each polynomial of the adjacency matrix. In
comparison, GCN and GraphSAGE do not improve much after
five layers, perhaps also suffering from oversmoothing.
2) Graph Convolution and Graph Pooling Comparison:
Among the pooling algorithms, DiffPool generally performs
the best. SagPool and SortPool perform better for MUTAG and
Proteins, but similar or worse for IMDB-Binary and Reddit-
Binary. Top-k pool performs poorly, suggesting that it requires
the auto-encoder structure to perform better. In general, only
Diffpool is consistently better than no pooling.
The results for graph convolution apply to graph pooling
with graph convolution. TAGCN with pooling generally per-
forms better than GCN and GraphSAGE with pooling and
more prone to overfitting, likely due to the same reasons.
V. CONCLUSION
On average, TAGCN generally performs well against GCN
and GraphSAGE on graph classification datasets with and
without pooling for sparser and larger graphs. We also find
that DiffPool generally outperforms the other pooling methods
evaluated. For future work, we would like to develop a better
theoretical understanding of GCNNs, by studying different
problems like oversmoothing and the design of different
parameters.
REFERENCES
[1] M. M. Bronstein, J. Bruna, Y. LeCun, A. Szlam, and P. Vandergheynst,
“Geometric Deep Learning: Going Beyond Euclidean Data,” IEEE
Signal Processing Magazine, vol. 34, no. 4, pp. 18–42, July 2017.
[2] A. Sandryhaila and J. M. F. Moura, “Discrete Signal Processing on
Graphs,” IEEE Trans. Signal Proc., vol. 61, no. 7, pp. 1644–1656, April
2013.
[3] A. Ortega, P. Frossard, J. Kovaevi, J. M. F. Moura, and P. Vandergheynst,
“Graph Signal Processing: Overview, Challenges, and Applications,”
Proceedings of the IEEE, vol. 106, no. 5, pp. 808–828, May 2018.
[4] J. Bruna, W. Zaremba, A. Szlam, and Y. LeCun, “Spectral Networks and
Locally Connected Networks on Graphs,” in International Conference
on Learning Representations (ICLR), 2014.
[5] M. Defferrard, X. Bresson, and P. Vandergheynst, “Convolutional Neural
Networks on Graphs with Fast Localized Spectral Filtering,” in Advances
in Neural Information Processing Systems (NIPS), 2016.
[6] T. N. Kipf and M. Welling, “Semi-Supervised Classification with Graph
Convolutional Networks,” in 5th International Conference on Learning
Representations, (ICLR) 2017, Toulon, France, dApril 24-26, 2017,
Conference Track Proceedings, 2017.
[7] J. Du, S. Zhang, G. Wu, J. M. F. Moura, and S. Kar, “Topology Adaptive
Graph Convolutional Networks,” Computing Research Repository, vol.
abs/1710.10370, 2017. [Online]. Available: http://arxiv.org/abs/1710.
10370
[8] J. Du, J. Shi, S. Kar, and J. M. F. Moura, “On Graph Convolution For
Graph CNNs,” in 2018 IEEE Data Science Workshop (DSW), June 2018,
pp. 1–5.
[9] J. Shi, M. Cheung, J. Du, and J. M. F. Moura, “Classification with
Vertex-Based Graph Convolutional Neural Networks,” in 2018 52nd
Asilomar Conference on Signals, Systems, and Computers, Oct 2018,
pp. 752–756.
[10] W. Hamilton, Z. Ying, and J. Leskovec, “Inductive Representation
Learning on Large Graphs,” in Advances in Neural
Information
Processing Systems 30, I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach,
R. Fergus, S. Vishwanathan, and R. Garnett, Eds. Curran Associates,
Inc., 2017, pp. 1024–1034. [Online]. Available: http://papers.nips.cc/
paper/6703-inductive-representation-learning-on-large-graphs.pdf
[11] P. Velikovi, G. Cucurull, A. Casanova, A. Romero, P. Li`o, and Y. Ben-
gio, “Graph Attention Networks,” in 6th International Conference on
Learning Representations (ICLR), 2018, Vancouver, BC, Canada, April
30 - May 3, 2018, Conference Track Proceedings, 2018.
[12] R. Ying, J. You, C. Morris, X. Ren, W. L. Hamilton, and J. Leskovec,
“Hierarchical Graph Representation Learning with Differentiable
Pooling,” in 32nd International Conference on Neural Information
Processing Systems (NIPS). Curran Associates Inc., 2018, pp. 4805–
4815. [Online]. Available: http://dl.acm.org/citation.cfm?id=3327345.
3327389
[13] H. Gao and S. Ji, “Graph U-Nets,” in 36th International Conference
on Machine Learning, ser. Proceedings of Machine Learning Research,
K. Chaudhuri and R. Salakhutdinov, Eds., vol. 97. Long Beach,
California, USA: PMLR, 09–15 Jun 2019, pp. 2083–2092. [Online].
Available: http://proceedings.mlr.press/v97/gao19a.html
[14] M. Zhang, Z. Cui, M. Neumann, and Y. Chen, “An End-to-End
Deep Learning Architecture for Graph Classification,” in 32nd
AAAI Conference on Artificial Intelligence, 2018. [Online]. Available:
https://aaai.org/ocs/index.php/AAAI/AAAI18/paper/view/17146
[15] S. Chen, R. Varma, A. Sandryhaila, and J. Kovaevi, “Discrete Signal
Processing on Graphs: Sampling Theory,” IEEE Transactions on Signal
Processing, vol. 63, no. 24, pp. 6510–6523, Dec 2015.
[16] A. Anis, A. Gadde, and A. Ortega, “Efficient Sampling Set Selection
for Bandlimited Graph Signals Using Graph Spectral Proxies,” IEEE
Transactions on Signal Processing, vol. 64, no. 14, pp. 3775–3789, July
2016.
[17] D. K. Hammond, P. Vandergheynst, and R. Gribonval, “Wavelets on
Graphs via Spectral Graph Theory,” Applied and Computational Har-
monic Analysis, vol. 30, no. 2, pp. 129 – 150, 2011. [Online]. Available:
http://www.sciencedirect.com/science/article/pii/S1063520310000552
[18] E. Luzhnica, B. Day, and P. Lio’, “Clique Pooling for Graph Classifi-
cation,” Computing Research Repository, vol. abs/1904.00374, 2019.
[19] Y. Boykov and O. Veksler, “Graph Cuts in Vision and Graphics:Theories
and Application,” in Handbook of Mathematical Models in Computer
Vision, N. Paragios, Y. Chen, and O. D. Faugeras, Eds. Springer, 2006,
pp. 79–96.
[20] N. Shervashidze, P. Schweitzer, E. J. van Leeuwen, K. Mehlhorn,
and K. M. Borgwardt, “Weisfeiler-Lehman Graph Kernels,” J. Mach.
Learn. Res., vol. 12, pp. 2539–2561, Nov. 2011. [Online]. Available:
http://dl.acm.org/citation.cfm?id=1953048.2078187
[21] J. Lee,
I. Lee, and J. Kang, “Self-Attention Graph Pooling,” in
Proceedings of the 36th International Conference on Machine Learning,
ser. Proceedings of Machine Learning Research, K. Chaudhuri and
R. Salakhutdinov, Eds., vol. 97.
Long Beach, California, USA:
PMLR, 09–15 Jun 2019, pp. 3734–3743.
[Online]. Available:
http://proceedings.mlr.press/v97/lee19c.html
[22] A. K. Debnath, R. L. Lopez de Compadre, G. Debnath, A. J. Shusterman,
and C. Hansch, “Structure-activity Relationship of Mutagenic Aromatic
and Heteroaromatic Nitro Compounds. Correlation with Molecular Or-
bital Energies and Hydrophobicity,” Journal of Medicinal Chemistry,
vol. 34, no. 2, pp. 786–797, 1991.
[23] B. Karsten, C. S. Ong, S. Schnauer, S. Vishwanathan, A. J. Smola,
and H.-P. Kriegel, “Protein Function Prediction via Graph Kernels,”
Intelligent Systems for Molecular Biology, 2005.
[24] P. Yanardag and S. Vishwanathan, “Deep Graph Kernels,” in Proceedings
of the 21th ACM SIGKDD International Conference on Knowledge
Discovery and Data Mining, ser. KDD ’15. New York, NY, USA:
ACM, 2015, pp. 1365–1374.
[25] M. Fey and J. E. Lenssen, “Fast Graph Representation Learning with
PyTorch Geometric,” in ICLR Workshop on Representation Learning on
Graphs and Manifolds, 2019.
�








 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc