Clustering and Projected Clustering with
Adaptive Neighbors
Feiping Nie
Department of Computer
Science and Engineering
University of Texas, Arlington
Texas, USA
feipingnie@gmail.com
xqwang1991@gmail.com
Texas, USA
Xiaoqian Wang
Department of Computer
Science and Engineering
Heng Huang∗
Department of Computer
Science and Engineering
University of Texas, Arlington
University of Texas, Arlington
Texas, USA
heng@uta.edu
ABSTRACT
Many clustering methods partition the data groups based
on the input data similarity matrix. Thus, the clustering
results highly depend on the data similarity learning. Be-
cause the similarity measurement and data clustering are
often conducted in two separated steps, the learned data
similarity may not be the optimal one for data clustering
and lead to the suboptimal results. In this paper, we pro-
pose a novel clustering model to learn the data similarity
matrix and clustering structure simultaneously. Our new
model learns the data similarity matrix by assigning the
adaptive and optimal neighbors for each data point based
on the local distances. Meanwhile, the new rank constraint
is imposed to the Laplacian matrix of the data similarity
matrix, such that the connected components in the resulted
similarity matrix are exactly equal to the cluster number.
We derive an efficient algorithm to optimize the proposed
challenging problem, and show the theoretical analysis on
the connections between our method and the K-means clus-
tering, and spectral clustering. We also further extend the
new clustering model for the projected clustering to han-
dle the high-dimensional data. Extensive empirical results
on both synthetic data and real-world benchmark data sets
show that our new clustering methods consistently outper-
forms the related clustering approaches.
Categories and Subject Descriptors
H.2.8 [Database Management]: Database Applications-
Data Mining
General Terms
Algorithms
Corresponding author.
Keywords
Clustering; Block diagonal similarity matrix; Adaptive neigh-
bors; Clustering with dimensionality reduction
∗
Permission to make digital or hard copies of all or part of this work for personal
or classroom use is granted without fee provided that copies are not made or
distributed for profit or commercial advantage and that copies bear this notice
and the full citation on the first page. Copyrights for components of this work
owned by others than ACM must be honored. Abstracting with credit is
permitted. To copy otherwise, or republish, to post on servers or to redistribute
to lists, requires prior specific permission and/or a fee. Request permissions
from permissions@acm.org.
KDD’14, August 24–27, 2014, New York, New York, USA.
Copyright © 2014 ACM 978-1-4503-2956-9/14/08…$15.00.
http://dx.doi.org/10.1145/2623330.2623726 .
1.
INTRODUCTION
Clustering, which partitions the data points into different
groups such that the objects in the same group have high
similarity to each other, is one of the most fundamental topic
in data mining. The clustering technique has been playing
an outstanding role in data mining applications. In the past
decades, many clustering algorithms have been proposed,
such as hierarchical clustering [10], K-means clustering [13],
spectral clustering [15], spectral embedded clustering [19],
support vector clustering [1], maximum margin clustering
[22],
initialization independent clustering [18], multi-view
clustering [21, 3, 4], etc.
Due to the efficiency and simpleness, the most popularly
used clustering method is the K-means clustering algorithm,
which aims to learn c cluster centroids that minimize the
within cluster data distances. The spectral clustering method
[15] does a low-dimension embedding of the affinity matrix
between samples, followed by a K-means clustering in the
low dimensional space. Because the data graph and manifold
information are utilized in the clustering model, the graph
based clustering methods (such as normalized cut [20], ra-
tio cut [7]) usually show better clustering performance than
K-means method. Such methods especially work well for
a small number of clusters. More recently, the Nonnega-
tive Matrix Factorization (NMF) has been used as the re-
laxation technique for clustering with excellent performance
[12, 16]. Although the graph-based clustering methods have
good performance, they partition the data based on the fixed
data graph such that the clustering results are sensitive to
the input affinity matrix.
In this paper, we propose to solve the clustering prob-
lem from a new point of view and learn the data similar-
ity matrix by assigning the adaptive and optimal neighbors
for each data point based on the local connectivity. Our
main assumption is that the data points with a smaller dis-
tance should have a larger probability to be neighbors. More
important, we impose the rank constraint on the Lapla-
cian matrix of the learned similarity matrix to achieve the
ideal neighbors assignment, such that the connected com-
ponents in the data are exact the cluster number and each
connected component corresponds to one cluster. Our new
model learns the data similarity matrix and cluster struc-
ture simultaneously to achieve the optimal clustering results.
We derive a novel and efficient algorithm to solve this chal-
lenging problem, and show the theoretical analysis on the
connections between our method and K-means clustering,
and spectral clustering. Moreover, we extend the proposed
977�
clustering model for the projected clustering to handle the
high-dimensional data. Extensive experiments have been
conducted on both synthetic data and real-world benchmark
data sets. All empirical results show that our new clustering
models consistently outperform the related clustering meth-
ods.
Notations: Throughout the paper, all the matrices are
written as uppercase. For matrix M , the i-th row (with
transpose) and the (i, j)-th element of M are denoted by mi
and mij, respectively. The trace of matrix M is denoted
by T r(M ). The L2-norm of vector v is denoted by v2,
the Frobenius norm of matrix M is denoted by MF . An
identity matrix is denoted by I, and 1 denotes a column
vector with all the elements as one. For vector v and matrix
M , v ≥ 0 and M ≥ 0 mean all the elements of M and v are
equal to or larger than zero.
2. CLUSTERING WITH ADAPTIVE NEIGH-
BORS
Exploring the local connectivity of data is a successful
strategy for clustering task. Given a data set {x1, x2, ..., xn},
we denote X ∈ R
n×d as the data matrix. The neighbors of
xi ∈ R
d×1 can be defined as the k-nearest data points in the
data set to xi. In this paper, we consider the probabilistic
neighbors, and use the Euclidean distance as the distance
measure for simplicity.
For the i-th data point xi, all the data points {x1, x2, ..., xn}
can be connected to xi as a neighbor with probability sij .
Usually, a smaller distance xi − xj2
2 should be assigned a
larger probability sij, so a natural method to determine the
probabilities sij|n
j=1 is solving the following problem:
min
i 1=1,0≤si≤1
sT
j=1
xi − xj2
2 sij
(1)
where si ∈ R
n×1 is a vector with the j-th element as sij.
However, the problem (1) has a trivial solution, only the
nearest data point can be the neighbor of xi with probability
1 and all the other data points can not be the neighbors of
xi. On the other hand, if we solve the following problem
without involving any distance information in the data:
n
n
min
i 1=1,0≤si≤1
sT
j=1
2
ij
s
(2)
the optimal solution is that all the data points can be the
neighbors of xi with the same probability 1
n , which can be
seen as a prior in the neighbors assignment.
Combining Eq.(1) and (2), we can solve the following
problem:
n
xi − xj2
2 sij + γs
2
ij
(3)
min
i 1=1,0≤si≤1
sT
j=1
The second term in Eq.(3) is a regularization and γ is the
regularization parameter. Denote dx
2, and de-
n×1 as a vector with the j-th element as dx
note dx
ij ,
then the problem (3) can be written in vector form as
ij = xi − xj2
i ∈ R
si +
2
2
1
2γ
x
i
d
min
i 1=1,0≤si≤1
sT
(4)
For each data point xi, we can use Eq.(3) to assign its
neighbors. Therefore, we can solve the following problem to
assign the neighbors for all the data points:
n
xi − xj2
2 sij + γs
2
ij
(5)
min
∀i,sT
i 1=1,0≤si≤1
i,j=1
n
In the clustering task to partition the data into c clusters,
an ideal neighbors assignment is that the connected com-
ponents in the data are exact c. Usually the neighbors as-
signment with Eq.(5) can not reach the ideal case for any
value of γ. In most cases, all the data points are connected
as just one connected component. In order to achieve the
ideal neighbors assignment, the probabilities sij|n
i,j=1 in the
problem (5) should be constrained such that the neighbors
assignment becomes an adaptive process to make the con-
nected components are exact c.
It seems look like an im-
possible goal since this kind of structured constraint on the
similarities is fundamental but also very difficult to han-
dle. In this paper, we will propose a novel but very simple
method to achieve this goal.
n×n obtained in the neighbors assign-
ment can be seen as a similarity matrix of the graph with
the n data points as the nodes. Suppose each node i is as-
signed a function value as fi ∈ R
c×1, then it can be verified
that
The matrix S ∈ R
fi − fj2
2 sij = 2T r(F
T
LSF )
(6)
i,j=1
where F ∈ R
DS − ST +S
degree matrix DS ∈ R
where the i-th diagonal element is
n×c with the i-th row formed by fi, LS =
is called Laplacian matrix in graph theory, the
n×n is defined as a diagonal matrix
j (sij + sji)/2.
2
If the similarity matrix S is nonnegative, then the Lapla-
cian matrix has an important property as follows [14, 5].
Theorem 1. The multiplicity c of the eigenvalue 0 of the
Laplacian matrix LS is equal to the number of connected
components in the graph with the similarity matrix S.
Theorem 1 indicates that if rank(LS) = n − c, then the
neighbors assignment is an ideal assignment and we already
partition the data points into c clusters based on S, without
having to perform K-means or other discretization proce-
dures. Motivated by Theorem 1, we add an additional con-
straint rank(LS) = n− c into the problem (5) to achieve the
ideal neighbors assignment with clear clustering structure.
Thus, our new clustering model is to solve:
xi − xj2
n
i 1 = 1, 0 ≤ si ≤ 1, rank(LS) = n − c
2 sij + γs2
i,j=1
ij
Jopt = min
s.t. ∀i, sT
S
(7)
2
ST +S
It is difficult to solve the problem (7). Because LS = DS−
and DS also depends on S, the constraint rank(LS) =
n − k is not easy to tackle. In the next subsection, we will
propose a novel and efficient algorithm to solve this chal-
lenging problem.
2.1 Optimization Algorithm Solving Problem
(7)
We will see in Subsection 2.4 that this problem can be solved
with a closed form solution.
Suppose σi(LS) is the i-th smallest eigenvalue of LS, we
know σi(LS) ≥ 0 since LS is positive semi-definite. It can
978�
c
ij
+ 2λ
σi(LS)
(8)
i=1
be seen that the problem (7) is equivalent to the following
problem for a large enough value of λ:
S
i,j=1
xi − xj2
n
2 sij + γs2
min
i 1 = 1, 0 ≤ si ≤ 1
s.t. ∀i, sT
c
c
When λ is large enough, note that σi(LS) ≥ 0 for every i,
then the optimal solution S to the problem (8) will make the
i=1 σi(LS) to be zero, and thus the constraint
second term
rank(LS) = n − c in the problem (7) could be satisfied.
According to the Ky Fan’s Theorem [6], we have
σi(LS) =
min
F∈Rn×c,F T F =I
i=1
T
T r(F
LSF )
(9)
Therefore, the problem (8) is further equivalent to the fol-
lowing problem
xi − xj2
n
min
S,F
s.t. ∀i, sT
i,j=1
2 sij + γs2
+ 2λT r(F T LSF )
i 1 = 1, 0 ≤ si ≤ 1, F ∈ Rn×c, F T F = I
ij
(10)
Compared with the original problem (7), the problem (10)
is much easier to solve. We can apply the alternative opti-
mization approach to solve it.
When S is fixed, the problem (10) becomes
min
F∈Rn×c,F T F =I
T
T r(F
LSF )
(11)
The optimal solution F to the problem (11) is formed by the
c eigenvectors of LS corresponding to the c smallest eigen-
values.
When F is fixed, the problem (10) becomes
+ 2λT r(F T LSF )
S
i,j=1
n
min
s.t. ∀i, sT
n
min
s.t. ∀i, sT
i,j=1
S
xi − xj2
2 sij + γs2
i 1 = 1, 0 ≤ si ≤ 1
ij
xi − xj2
2 sij + γs2
i 1 = 1, 0 ≤ si ≤ 1
According to Eq.(6), the problem (12) can be rewritten as
ij + λ fi − fj2
2 sij
(13)
(14)
(15)
Note that the problem (13) is independent between different
i, so we can solve the following problem individually for each
i:
2 sij + γs2
ij + λ fi − fj2
2 sij
xi − xj2
n
j=1
i 1 = 1, 0 ≤ si ≤ 1
sT
ij = xi − xj2
min
si
s.t.
n×1 as a vector with the j-th element as dij = dx
2, and denote
ij +
f
ij , then the problem (14) can be written in vector form
2 and d
f
ij = fi − fj2
Denote dx
di ∈ R
λd
as
si +
2
2
1
2γ
di
min
i 1=1,0≤si≤1
sT
We will see in Subsection 2.4 that this problem can be solved
with a closed form solution.
The detailed algorithm to solve the problem (7) is sum-
marized in Algorithm 1.
1In practice, to accelerate the procedure, the λ can be de-
termined during the iteration. We can initialize λ = γ, then
increase λ if the connected components of S is smaller than
c and decrease λ if it is greater than c in each iteration.
n×d, cluster number c, parameter
Algorithm 1 Algorithm to solve problem (7).
input Data matrix X ∈ R
output S ∈ R
γ, a large enough λ1.
n×n with exact c connected components.
Initialize S by the optimal solution to the problem (5).
while not converge do
1. Update F , which is formed by the c eigenvectors
of LS = DS − ST +S
corresponding to the c smallest
eigenvalues.
2. For each i, update the i-th row of S by solving the
problem (15), where di ∈ R
n×1 is a vector with the j-th
element as dij = xi − xj2
2 + λ fi − fj2
2.
2
end while
2.2 Connection to K-means Clustering
Denote the centering matrix by
H = I − 1
n
T
11
(16)
and denote Dx ∈ R
n×n as a distance matrix where the (i, j)-
th element is dx
2. To analyze the connection of
Algorithm 1 to K-means, we first need the following lemma:
ij = xi − xj2
,
Lemma 1. HDxH = −2HXX T H
2 = xT
ij = xi − xj2
i xi + xT
Proof. Since dx
i xj, we
have Dx = Diag(XX T )11T + 11T Diag(XX T ) − 2XX T ,
where Diag(XX T ) is a diagonal matrix with the diagonal
elements of XX T . Note that H1 = 1T H = 0 according to
the definition of H, we have HDxH = −2HXX T H.
2
Theorem 2 reveals that Algorithm 1 is exactly solving the
K-means problem when γ → ∞.
j xj − 2xT
to the problem of K-means.
Proof. The problem (7) can be written in matrix form as
T
x
) + γ S2
F
D
min
T r(S
S1=1,S≥0,rank(LS )=n−c
(17)
Due to the constraint rank(LS) = n − c, the solution S
has exact c components (that is, S is block diagonal with
proper permutation). Suppose the i-th component of S is
Si ∈ R
ni×ni , where ni is the number of data points in the
component, then solving problem (17) is to solve the follow-
ing problem for each i:
i ) + γ Si2
When γ → ∞, then the problem (18) becomes
Si1=1,Si≥0
T r(S
T
i D
min
F
x
(18)
(19)
min
Si1=1,Si≥0
Si2
F
The optimal solution to the problem (19) is that all the
elements of Si are equal to 1
ni
should be the following form when γ → ∞:
Therefore, the optimal solution S to the problem (17)
.
sij =
1
nk
0
xi, xj are in the same component k
otherwise
(20)
We denote the solution set that satisfies the form in Eq.(20)
by V. Note that for any possible partition of the c com-
ponents such that S has the form as in Eq.(20), S2
F has
(12)
Theorem 2. When γ → ∞, the problem (7) is equivalent
979�
the same value, i.e., S2
becomes
F = c. Therefore, the problem (17)
S∈V T r(S
min
T
x
)
D
(21)
According to the constraint of S in Eq.(21), S is symmet-
ric and S1 = 1T S = 1. So T r(HDxHS) = T r(DxS) −
n 1T Dx1 and thus the problem (21) is equivalent to the fol-
lowing problem:
1
S∈V T r(HD
min
Define the label matrix Y ∈ R
ment is
1√
x
HS)
(22)
n×c, where the (i, j)-th ele-
yij =
nk
0
xi belongs to the k-th compoment
otherwise
Then according to Eq.(22) and Lemma 1, we have
S∈V T r(HXX T HS)
S∈V T r(X T HSHX)
S∈V T r(X T H(I − S)HX)
S∈V T r(HDxHS)
min
⇔ max
⇔ max
⇔ min
⇔ min
⇔ min
T r(X T H(I − Y Y T )HX)
T r(Sw)
Y
Y
(23)
(24)
which is exactly the problem of K-means. The matrix Sw is
called within-class scatter matrix in classical Linear Discrim-
inant Analysis (LDA) when the labels Y of data are given.
K-means is to find the optimal labels Y such that the trace
of the within-class scatter matrix T r(Sw) is minimized. 2
We will see in the next subsection that the proposed method
in Algorithm 1 is closely related to spectral clustering. There-
fore, although the new algorithm is to solve the K-means
problem (which can only partition data with spherical shape)
when γ → ∞, it can partition data with arbitrary shape
when γ is not very large. We will also see in the experiments
that this new algorithm can find much better solution to the
K-means problem even when γ is not very large.
2.3 Connection to Spectral Clustering
Given a graph with the similarity S, spectral clustering is
to solve the following problem:
min
F∈Rn×c,F T F =I
T
T r(F
LSF )
(25)
The optimal solution is the spectral decomposition of the
Laplacian matrix LS, i.e., the optimal solution F is formed
by the c eigenvectors of LS corresponding to the c smallest
eigenvalues, as in Eq.(11).
Usually, given a similarity S, the obtained F can not be
directly used for clustering since the graph with S does not
has exact c connected components. K-means or other dis-
cretization procedures should be performed on F to obtain
the final clustering results [9].
In the convergence of Algorithm 1, we also obtain an opti-
mal solution F to the problem (25), the difference is that the
similarity S is also learned by the algorithm. Note that in
the convergence, the last term 2λT r(F T LSF ) in the problem
(10) will approximate zero, the learned S is mainly achieved
by solving problem (5).
Thanks to the constraint rank(LS) = n − c, the graph
with the learned S will has exact c connected components.
Therefore, the optimal solution F , which is formed by the c
eigenvectors of LS corresponding to the c smallest eigenval-
ues, can be written as
F = Y Q
(26)
where Y ∈ R
n×c is the label matrix defined in Eq.(23) and
Q ∈ R
c×c is an arbitrary orthogonal matrix. That is to
say, we can use the obtained F directly to get the final clus-
tering result, without the K-means or other discretization
procedures as traditional spectral clustering has to do.
In another viewpoint, it can be seen that the proposed
algorithm learns the similarity matrix S and the label matrix
F simultaneously, while traditional spectral clustering only
learns the F given the similarity matrix S. Therefore, the
new algorithm could achieve better performance in practice
since it also learns an adaptive graph for clustering.
2.4 Determine The Value of γ
In practice, regularization parameter is difficult to tune
since its value could be from zero to infinite. In this sub-
section, we present an effective method to determine the
regularization parameter γ in the problem (7).
For each i, the objective function in the problem (7) is
equal to the one in the problem (4). The Lagrangian func-
tion of problem (4) is
L(si, η, βi) =
T
i si
where η and βi ≥ 0 are the Lagrangian multipliers.
T
2
− η(s
i 1 − 1) − β
dx
i
2γi
1
2
(27)
si +
2
According to the KKT condition [2], it can be verified that
the optimal solution si should be
sij = ( − dx
ij
2γi
+ η)+
(28)
In practice, usually we could achieve better performance if
we focus on the locality of data. Therefore, it is preferred to
learn a sparse si, i.e., only the k nearest neighbors of xi could
have chance to connect to xi. Another benefit of learning a
sparse similarity matrix S is that the computation burden
can be alleviated largely for subsequent processing.
i2, ..., dx
in are or-
If the optimal si has only k
dered from small to large.
nonzero elements, then according to Eq.(28), we know sik >
0 and si,k+1 = 0. Therefore, we have
Without loss of generality, suppose dx
i1, dx
− dx
− dx
2γi + η ≤ 0
ik
2γi
i,k+1
+ η > 0
(29)
According to Eq.(28) and the constraint sT
i 1 = 1, we have
k
k
j=1
2γi + η) = 1
ij
j=1
( − dx
⇒ η = 1
k + 1
2kγi
k
j=1
dx
ij
(30)
So we have the following inequality for γi according to Eq.(29)
and Eq.(30):
x
ik − 1
2
d
k
2
x
ij < γi ≤ k
2
d
x
i,k+1 − 1
2
d
x
ij
d
(31)
k
j=1
Therefore, in order to obtain an optimal solution si to the
problem (4) that has exact k nonzero values, we could set
980�
γi to be
γi=
x
i,k+1 − 1
2
d
k
2
k
j=1
x
ij
d
(32)
The overall γ could be set to the mean of γ1, γ2, ..., γn. That
is, we could set the γ to be
k
γ=
1
n
x
i,k+1 − 1
2
d
k
2
x
ij
d
(33)
n
i=1
When F is fixed, the problem (36) becomes
n
W T xi − W T xj
2
2 sij + γs2
ij
i,j=1
+2λT r(F T LSF )
s.t. ∀i, sT
i 1 = 1, 0 ≤ si ≤ 1, W T StW = I
min
S,W,F
(37)
j=1
In problem (37), if S is fixed, then the problem becomes
The number of neighbors k is much easier to tune than the
regularization parameter γ since k is an integer and has ex-
plicit meaning.
3. PROJECTED CLUSTERING WITH
ADAPTIVE NEIGHBORS
Clustering high-dimensional data is an important and chal-
lenging problem in practice. In this section, we propose a
projected clustering approach with the adaptive neighbors
to solve this problem. The goal is to find an optimal sub-
space on which the adaptive neighboring is performed such
that there are exact c connected components in the data.
Denote the total scatter matrix by St = X T HX, where H
is the centering matrix defined in Eq.(16). Suppose we are
to learn a projection matrix W ∈ R
d×m. First, we constrain
the subspace with W T StW = I such that the data on the
subspace are statistically uncorrelated. As in Eq.(5), we
assign the neighbors for each data by solving the following
problem:
n
min
S,W
s.t. ∀i, sT
W T xi − W T xj
i 1 = 1, 0 ≤ si ≤ 1, W T StW = I
2 sij + γs2
2
i,j=1
ij
(34)
n
W
i,j=1
2
2
T
xi − W
T
xj
min
W T StW =I
sij
(38)
which can be rewritten as the following problem according
to Eq.(6):
min
W T StW =I
T r(W
T
T
X
LSXW )
(39)
The optimal solution W to the problem (39) is formed by
−1
t X T LSX corresponding to the m
the m eigenvectors of S
smallest eigenvalues (we assume the null space of the data
X is removed, i.e., St is invertible).
In problem (37), if W is fixed, then according to Eq.(6)
again, the problem (37) can be rewritten as
2
min
n
W T xi − W T xj
n
2 sij
i 1 = 1, 0 ≤ si ≤ 1
s.t. ∀i, sT
fi − fj2
+λ
i,j=1
i,j=1
S
2 sij + γs2
ij
(40)
Similarly, to make the neighbors assignment be adaptive
such that the connected components in the data are exact
c, we impose the constraint on S with rank(LS) = n − c.
Therefore, we propose the following problem for learning the
projection W and the clustering simultaneously:
n
min
S,W
s.t. ∀i, sT
W T xi − W T xj
i 1 = 1, 0 ≤ si ≤ 1, W T StW = I,
2 sij + γs2
2
i,j=1
ij
rank(LS) = n − c
3.1 Optimization Algorithm for Problem (35)
Using the same trick as in Subsection 2.1, we know that
solving problem (35) is equivalent to solving the following
problem
n
W T xi − W T xj
2
2 sij + γs2
ij
min
S,W,F
s.t. ∀i, sT
i 1 = 1, 0 ≤ si ≤ 1, W T StW = I,
i,j=1
+2λT r(F T LSF )
F ∈ Rn×c, F T F = I
(35)
min
si
s.t.
Note that the problem (40) is independent between different
i, so we can solve the following problem individually for each
i:
2 sij + γs2
ij
2
j=1
n
W T xi − W T xj
n
fi − fj2
2 sij
+λ
i 1 = 1, 0 ≤ si ≤ 1
2
W T xi − W T xj
sT
j=1
(41)
Denote dwx
ij =
i ∈ R
denote dw
dw
ij = dwx
ij + λd
vector form as
2, and
n×1 as a vector with the j-th element as
f
ij , then the problem (41) can be written in
2 and d
f
ij = fi − fj2
(36)
min
i 1=1,0≤si≤1
sT
si +
2
2
1
2γ
w
i
d
(42)
We can also apply the alternative optimization approach to
solve this problem.
When S is fixed, the problem (36) becomes the problem
(11), and the optimal solution F is formed by the c eigen-
vectors of LS corresponding to the c smallest eigenvalues.
which is the same problem as in Eq.(15) and can be solved
with a closed form solution.
The detailed algorithm to solve the problem (35) is sum-
marized in Algorithm 2. We can also use Eq.(33) to deter-
mine the regularization parameter γ.
981�
Algorithm 2 Algorithm to solve problem (35).
input Data matrix X ∈ R
dimension m, parameter γ, a large enough λ.
output S ∈ R
jection W ∈ R
Initialize S by the optimal solution to the problem (3).
while not converge do
n×d, cluster number c, reduced
n×n with exact c connected components, pro-
d×m.
1. Update LS = DS − ST +S
n×n is
a diagonal matrix with the i-th diagonal element as
, where DS ∈ R
2
j(sij + sji)/2.
2. Update F , whose columns are the c eigenvectors of
LS corresponding to the c smallest eigenvalues.
2. Update W , whose columns are the m eigenvectors of
−1
t X T LSX corresponding to the m smallest eigenval-
S
ues.
3. For each i, update the i-th row of S by solving the
n×1 is a vector with the j-th
problem (42), where dw
element as dw
W T xi − W T xj
2
2 + λfi − fj2
2.
i ∈ R
ij =
end while
3.2 Connection to LDA
In this subsection, we show that when γ → ∞, the pro-
posed method in Algorithm 2 is to solve the LDA problem
with the labels also being optimized at the same time. The
result is summarized in the following theorem:
Theorem 3. When γ → ∞, the problem (35) is equiv-
alent to the problem of LDA, in which the labels are also
variables to be optimized.
Proof. The problem (35) can be written in matrix form as
Then according to Eq.(46) and Lemma 1, we have
T r(HDwxHS)
min
S∈V,W T StW =I
=
max
max
S∈V,W T StW =I
S∈V,W T StW =I
S∈V,W T StW =I
min
min
Y,W T StW =I
min
Y,W T StW =I
=
=
=
=
T r(HXW W T X T HS)
T r(W T X T HSHXW )
T r(W T X T H(I − S)HXW )
T r(W T X T H(I − Y Y T )HXW )
T r(W T SwW )
(47)
where Y is the label matrix defined as in Eq.(23).
If the
label matrix Y is given, then the optimal solution to the
problem (47) is equal to the solution of LDA. Therefore,
when γ → ∞, the proposed method in Algorithm 2 is to
solve the LDA problem, but the labels are also unknown
2
and are to be optimized at the same time.
When γ is not very large, the matrix X T LSX in Eq.(39)
can be viewed as local within-class scatter matrix. In this
case, the Algorithm 2 can be viewed as a unsupervised ver-
sion of local scatter matrices based LDA methods, which are
designed for handling multimodal non-Gaussian data [17].
4. EXPERIMENTS
In this part, we will show the performance of the proposed
clustering method (Algorithm 1) and the projected cluster-
ing method (Algorithm 2) on both synthetic data and real
data sets. For simplicity, we denote our clustering method
as CAN (Clustering with Adaptive Neighbors), and our pro-
jected clustering method as PCAN (Projected Clustering
with Adaptive Neighbors) in the following context.
4.1 Experiments on Synthetic Data
min
S1=1,S≥0,W T StW =I,
rank(LS )=n−c
T r(S
wx
T
D
) + γ S2
F
2
W T xi − W T xj
where Dwx ∈ R
n×n is a distance matrix with the (i, j)-th
element as dwx
ij =
2. Due to the constraint
rank(LS) = n − c, the solution S has exact c components.
Suppose the i-th component of S is Si ∈ R
ni×ni , where ni
is the number of data points in the component, then solving
problem (43) is to solve the following problem for each i:
Si1=1,Si≥0,W T StW =I
min
T r(S
T
wx
i D
i
) + γ Si2
F
(44)
When γ → ∞, then the problem (44) becomes
min
Si1=1,Si≥0
Si2
F
(45)
The optimal solution to the problem (45) is that all the
elements of Si are equal to 1
ni .
With similar derivation as in Subsection 2.2, we know the
problem (43) is equivalent to the following problem when
γ → ∞:
min
S∈V,W T StW =I
(46)
where V is the solution set of S that satisfies the form in
Eq.(20).
T r(HD
HS)
wx
(43)
4.1.1 Experiments to Evaluate CAN Method
We use two synthetic data sets to measure the clustering
efficiency of CAN. Also, we find that CAN is able to obtain
an excellent initialization index vector for K-Means, which
decreases its objective value and promotes its clustering ac-
curacy to a large extent.
The first toy data set we used is a randomly generated
two-moon data. In this data set, there are two clusters of
data distributed in the two-moon shape. Our goal is to
construct a new similarity matrix to divide the data points
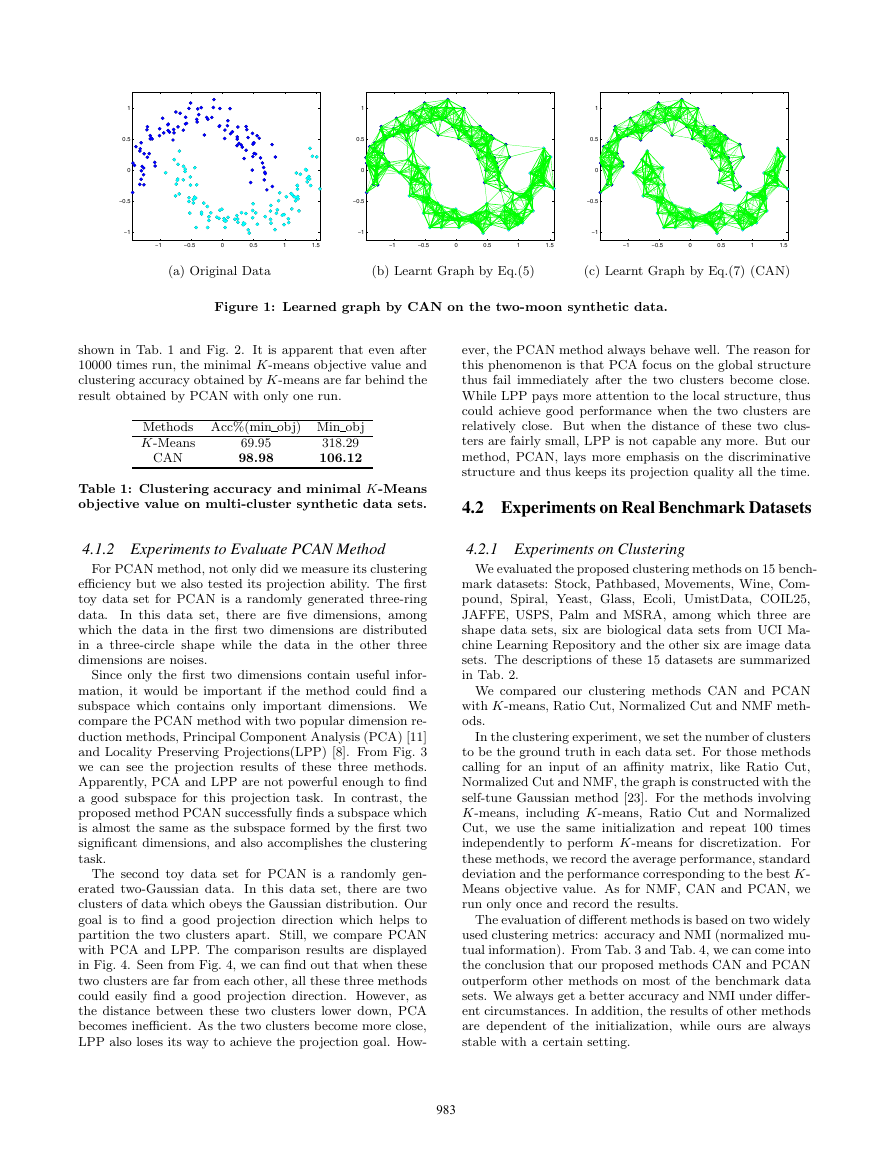
into exact two clusters. From Fig. 1 we can easily find the
clustering efficiency of the proposed CAN method. In this
figure, we set the color of the two clusters to be cyan and blue
separately and let the width of the connecting line denote
the learned similarity of two corresponding points. In the
initial similarity matrix learned by Eq.(5), several pairs of
points from different clusters are connected. Whereas, after
the learning by Eq.(7), there is not even a single line between
these two clusters, which means that the proposed clustering
method CAN can successfully partition the original data into
two classes.
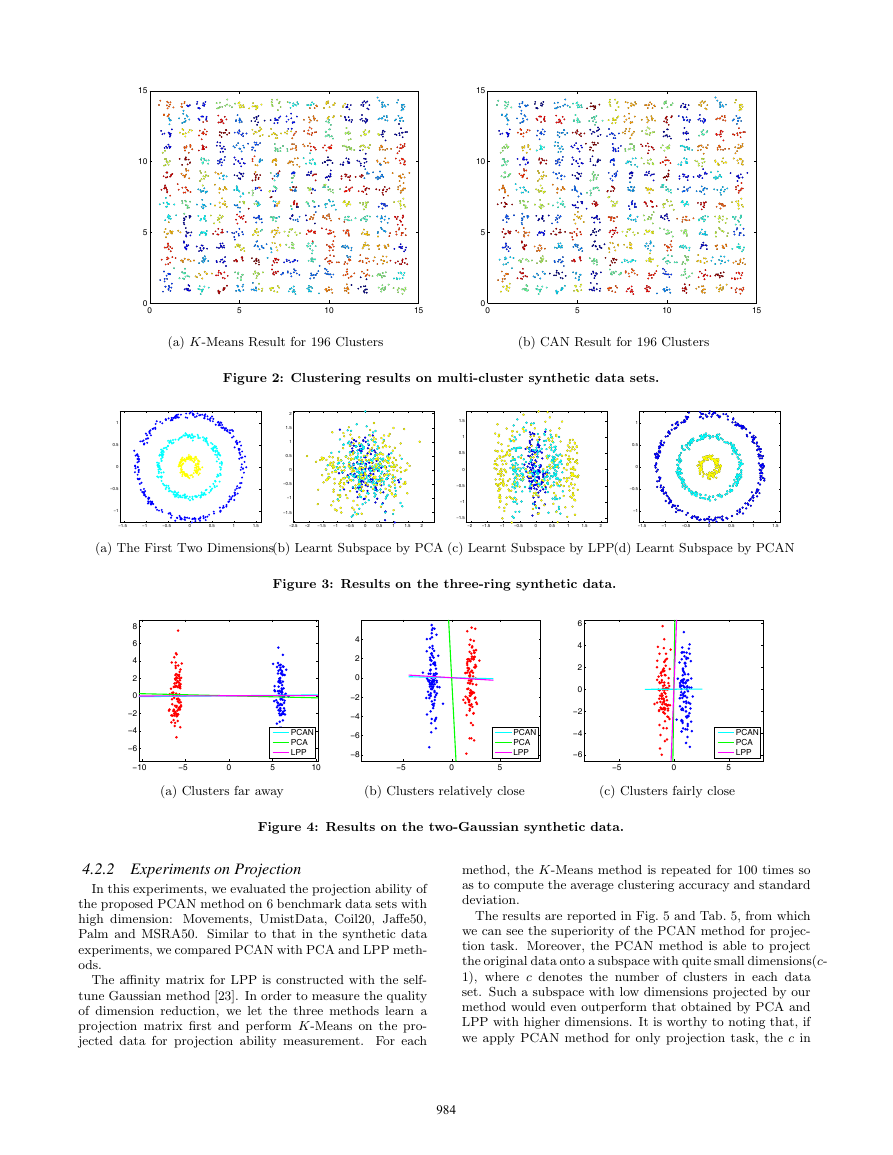
The second toy data set is a randomly generated multi-
cluster data.
In this data set, there are 196 clusters dis-
tributed in a spherical way. Firstly we run K-Means for
10000 times and report the result with the minimal K-means
objective value in the independently 10000 times run. Then
we run CAN once to generate an clustering result and use
it as initialization for K-means. The comparison results are
982�
1
0.5
0
−0.5
−1
1
0.5
0
−0.5
−1
1
0.5
0
−0.5
−1
−1
−0.5
0
0.5
1
1.5
−1
−0.5
0
0.5
1
1.5
−1
−0.5
0
0.5
1
1.5
(a) Original Data
(b) Learnt Graph by Eq.(5)
(c) Learnt Graph by Eq.(7) (CAN)
Figure 1: Learned graph by CAN on the two-moon synthetic data.
shown in Tab. 1 and Fig. 2. It is apparent that even after
10000 times run, the minimal K-means objective value and
clustering accuracy obtained by K-means are far behind the
result obtained by PCAN with only one run.
Methods Acc%(min obj) Min obj
K-Means
318.29
106.12
69.95
98.98
CAN
Table 1: Clustering accuracy and minimal K-Means
objective value on multi-cluster synthetic data sets.
ever, the PCAN method always behave well. The reason for
this phenomenon is that PCA focus on the global structure
thus fail immediately after the two clusters become close.
While LPP pays more attention to the local structure, thus
could achieve good performance when the two clusters are
relatively close. But when the distance of these two clus-
ters are fairly small, LPP is not capable any more. But our
method, PCAN, lays more emphasis on the discriminative
structure and thus keeps its projection quality all the time.
4.2 Experiments on Real Benchmark Datasets
4.1.2 Experiments to Evaluate PCAN Method
4.2.1 Experiments on Clustering
For PCAN method, not only did we measure its clustering
efficiency but we also tested its projection ability. The first
toy data set for PCAN is a randomly generated three-ring
data.
In this data set, there are five dimensions, among
which the data in the first two dimensions are distributed
in a three-circle shape while the data in the other three
dimensions are noises.
Since only the first two dimensions contain useful infor-
mation, it would be important if the method could find a
subspace which contains only important dimensions. We
compare the PCAN method with two popular dimension re-
duction methods, Principal Component Analysis (PCA) [11]
and Locality Preserving Projections(LPP) [8]. From Fig. 3
we can see the projection results of these three methods.
Apparently, PCA and LPP are not powerful enough to find
a good subspace for this projection task. In contrast, the
proposed method PCAN successfully finds a subspace which
is almost the same as the subspace formed by the first two
significant dimensions, and also accomplishes the clustering
task.
The second toy data set for PCAN is a randomly gen-
erated two-Gaussian data. In this data set, there are two
clusters of data which obeys the Gaussian distribution. Our
goal is to find a good projection direction which helps to
partition the two clusters apart. Still, we compare PCAN
with PCA and LPP. The comparison results are displayed
in Fig. 4. Seen from Fig. 4, we can find out that when these
two clusters are far from each other, all these three methods
could easily find a good projection direction. However, as
the distance between these two clusters lower down, PCA
becomes inefficient. As the two clusters become more close,
LPP also loses its way to achieve the projection goal. How-
We evaluated the proposed clustering methods on 15 bench-
mark datasets: Stock, Pathbased, Movements, Wine, Com-
pound, Spiral, Yeast, Glass, Ecoli, UmistData, COIL25,
JAFFE, USPS, Palm and MSRA, among which three are
shape data sets, six are biological data sets from UCI Ma-
chine Learning Repository and the other six are image data
sets. The descriptions of these 15 datasets are summarized
in Tab. 2.
We compared our clustering methods CAN and PCAN
with K-means, Ratio Cut, Normalized Cut and NMF meth-
ods.
In the clustering experiment, we set the number of clusters
to be the ground truth in each data set. For those methods
calling for an input of an affinity matrix, like Ratio Cut,
Normalized Cut and NMF, the graph is constructed with the
self-tune Gaussian method [23]. For the methods involving
K-means, including K-means, Ratio Cut and Normalized
Cut, we use the same initialization and repeat 100 times
independently to perform K-means for discretization. For
these methods, we record the average performance, standard
deviation and the performance corresponding to the best K-
Means objective value. As for NMF, CAN and PCAN, we
run only once and record the results.
The evaluation of different methods is based on two widely
used clustering metrics: accuracy and NMI (normalized mu-
tual information). From Tab. 3 and Tab. 4, we can come into
the conclusion that our proposed methods CAN and PCAN
outperform other methods on most of the benchmark data
sets. We always get a better accuracy and NMI under differ-
ent circumstances. In addition, the results of other methods
are dependent of the initialization, while ours are always
stable with a certain setting.
983�
15
10
5
0
0
1
0.5
0
−0.5
−1
15
10
5
0
0
5
10
15
5
10
15
(a) K-Means Result for 196 Clusters
(b) CAN Result for 196 Clusters
Figure 2: Clustering results on multi-cluster synthetic data sets.
2
1.5
1
0.5
0
−0.5
−1
−1.5
1.5
1
0.5
0
−0.5
−1
−1.5
1
0.5
0
−0.5
−1
−1.5
−1
−0.5
0
0.5
1
1.5
−2.5
−2
−1.5
−1
−0.5
0
0.5
1
1.5
2
−2
−1.5
−1
−0.5
0
0.5
1
1.5
2
−1.5
−1
−0.5
0
0.5
1
1.5
(a) The First Two Dimensions
(b) Learnt Subspace by PCA
(c) Learnt Subspace by LPP
(d) Learnt Subspace by PCAN
Figure 3: Results on the three-ring synthetic data.
8
6
4
2
0
−2
−4
−6
PCAN
PCA
LPP
−10
−5
0
5
10
4
2
0
−2
−4
−6
−8
PCAN
PCA
LPP
−5
0
5
6
4
2
0
−2
−4
−6
PCAN
PCA
LPP
−5
0
5
(a) Clusters far away
(b) Clusters relatively close
(c) Clusters fairly close
Figure 4: Results on the two-Gaussian synthetic data.
4.2.2 Experiments on Projection
In this experiments, we evaluated the projection ability of
the proposed PCAN method on 6 benchmark data sets with
high dimension: Movements, UmistData, Coil20, Jaffe50,
Palm and MSRA50. Similar to that in the synthetic data
experiments, we compared PCAN with PCA and LPP meth-
ods.
The affinity matrix for LPP is constructed with the self-
tune Gaussian method [23]. In order to measure the quality
of dimension reduction, we let the three methods learn a
projection matrix first and perform K-Means on the pro-
jected data for projection ability measurement. For each
method, the K-Means method is repeated for 100 times so
as to compute the average clustering accuracy and standard
deviation.
The results are reported in Fig. 5 and Tab. 5, from which
we can see the superiority of the PCAN method for projec-
tion task. Moreover, the PCAN method is able to project
the original data onto a subspace with quite small dimensions(c-
1), where c denotes the number of clusters in each data
set. Such a subspace with low dimensions projected by our
method would even outperform that obtained by PCA and
LPP with higher dimensions. It is worthy to noting that, if
we apply PCAN method for only projection task, the c in
984�














 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc