Agricultural Sciences, 2018, 9, 374-383
http://www.scirp.org/journal/as
ISSN Online: 2156-8561
ISSN Print: 2156-8553
Cloning and Expression of Anthocyanin
Biosynthesis Related Gene RrMYB6 in Rosa
rugosa
Kai Zou#, Yang Wang#, Mingyuan Zhao, Lanyong Zhao*, Zongda Xu*
Forestry College of Shandong Agricultural University, Tai’an, China
How to cite this paper: Zou, K., Wang, Y.,
Zhao, M.Y., Zhao, L.Y. and Xu, Z.D. (2018)
Cloning and Expression of Anthocyanin
Biosynthesis Related Gene RrMYB6 in Rosa
rugosa. Agricultural Sciences, 9, 374-383.
https://doi.org/10.4236/as.2018.93026
Received: January 30, 2018
Accepted: March 24, 2018
Published: March 27, 2018
Copyright © 2018 by authors and
Scientific Research Publishing Inc.
This work is licensed under the Creative
Commons Attribution International
License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/
Open Access
Abstract
R2R3-MYB transcription factor plays an important role in plant anthocyanin
synthesis. Based on the transcriptional database of Rosa rugosa, one MYB
transcription factor related to floral color, RrMYB6, was cloned. By using
bioinformatics analysis method, cloning MYB gene and analyzing its function
in anthocyanin biosynthesis regulation, we hope to lay a solid foundation for
new color variety breeding of R. rugosa. Using the R. rugosa “Zi zhi” as the
material, we obtained the total length of cDNA of RrMYB6 by RT-PCR and
RACE. By analyzing its bioinformatics, we found that the formula of the pro-
tein was C1491H2368N452O470S17, molecular weight was 34690.97 Da, the theoret-
ical pI was 8.74. In addition, it belonged to unstable protein with an unstable
index at 50.59, and it was also a hydrophilic protein with the total average hy-
drophobic index at −0.847. In the secondary structure of RrMYB6 protein, the
Alpha helix accounted for 32.35%, random coil was 47.39%, extended strand
was 11.11%, and beta turn was 9.15%. The sequence analysis showed that
RrMYB6 had a typical R2R3-MYB domain and bHLH binding domain, and it
also had an N1, C1, C2 inhibitory motif, belonging to the Sg4 subfamily MYB
protein. What’s more, evolutionary analysis indicated that the RrMYB6 pro-
tein was closely related with the MYB protein in Rosacea family, while it was
far from those in other families. The expression analysis showed that RrMYB6
protein decreased with the color of petals deeping, and its expression was the
lowest in the petals while the highest in stamens. According to the above re-
sults, it was speculated that RrMYB6 was involved in regulating the anthocya-
nin synthesis of R. rugosa, which belonged to negative regulatory mechanism.
Keywords
Rosa rugosa, Anthocyanin, R2R3-MYB, Gene Expression
#These authors contribute equally.
DOI: 10.4236/as.2018.93026 Mar. 27, 2018
374
Agricultural Sciences
�
K. Zou et al.
1. Introduction
Rosa rugosa, as a deciduous shrub in the Rosacea family, has high ornamental
value and fragrant aroma, and plays an important role in landscape architecture.
But the color of R. rugosa is simple, most is red, pink and white, and is fresh in
other color, which have seriously restricted its application in gardens for a long
time. In higher plants, the color of petals, tissues and fruits is determined by an-
thocyanin, which is a secondary metabolite belonging to flavonoids. Anthocya-
nins are synthesized through three steps with a series of enzymes [1]. The an-
thocyanins pathway has been explored in depth in Petunia hybrid [2], Zea mays
[3], Antirrhinum majus [4] and so on. In higher plants, most anthocyanin bio-
synthesis is regulated by the combination of many transcription factors in dif-
ferent time and space. TFs like R2R3-MYB, bHLH, WD-40 can regulate antho-
cyanin biosynthesis by forming a MBW ternary complex [5] [6]. As a key tran-
scription factor, the up-regulation of R2R3-MYB can activate the expression of
one or more structural genes involved in anthocyanin synthesis, so leads to an-
thocyanin accumulating. The related genes have been cloned and analyzed in its
function from species such as Epimedium sagittatum [7], Pyrus pyrifolia [8] and
Prunus avium L. [9]. At present, there are few reports on the anthocyanin bio-
synthesis mechanism of R. rugose, so we don’t know exactly how it works. Based
on the rose transcriptome data, we cloned one R2R3-MYB gene related with an-
thocyanin synthesis and figured out its function by bioinformatics analysis and
real-time PCR to provide foundation for the regulation mechanism of anthocya-
nin synthesis in the petals of R. rugosa.
2. Materials and Methods
The experiment was conducted from April 2016 to January 2017 at the flower
germplasm resource nursery of Shandong Agricultural University and the
Flower Institute of Forestry College.
2.1. Plant Materials
The plant material, Chinese representative R. rugosa “Zi zhi”, “Bai zizhi”, “Fen
zizhi” were from the rose germplasm resources garden at Shandong Agricultural
College. R. rugosa “Zi zhi”, “Bai zizhi”, “Fen zizhi” are the most representative
traditional rose in China. And the colors of three are purple, white and pink, re-
spectively. From April to May, 2016, the petals in initial opening stage of a single
plant with strong growth potential and stable hues in above three cultivars were
collected for the experiment of gene cloning and difference expression; Sepals,
stems, leaves, stamens, pistils and petals samples for gene spatial expression
analysis were collected from R. rugosa “Zi zhi” at full opening stage. All samples
were collected directly frozen with liquid nitrogen, and finally stored at −80˚C
until used.
375
Agricultural Sciences
DOI: 10.4236/as.2018.93026
�
K. Zou et al.
2.2. Methods
2.2.1. RNA Extraction and Gene Cloning
The extraction of RNA from various tissues of plants was operated according to
the instructions of EASYspin plant RNA rapid extraction kit, and its concentra-
tion and purity were determined by ultraviolet spectrophotometer. At the same
time, the integrity of RNA was detected by 1% agarose gel electrophoresis.
cDNA was synthesized by RNA reverse transcription kit according to the de-
scription of abm reverse transcription kit. Fragment amplification: according to
the data of rose transcriptome, the relevant MYB gene fragments were selected,
and specific primers were designed by Oligo 7.0 software (Table 1). PCR ampli-
fication was carried out with the synthesized cDNA as the template. The reaction
system of PCR was as follows: Mix 12.5 ul, the target gene upstream and down-
stream primers each 1 ul, template cDNA 1 ul, sterilization ddH2O supplement
to 25 ul; The PCR reaction conditions are as follows: 94˚C for 5 min; 94˚C for 30
s, 53˚C for 30 s, and 72˚C for 1 min for a total of 35 cycles; and then extension at
72˚C for 10 min. Next, 1% agarose gel electrophoresis was used to detect the
PCR products. The full-length amplification was carried out at 72˚C for 10 min.
Specific primers (Table 1) were used to amplify the three races and the complete
open reading frame (ORF). The reaction system and PCR reaction conditions
were described above. The PCR product was reclaimed according to the descrip-
tion of Hipure Gel Pure DNA Mini Kit(Magen), then ligated with the carrier
pMD18-T to transform Escherichia coli DH5α. Positive clones were selected and
sequenced at BGI.
2.2.2. Bioinformatics Analysis of RrMYB6
BLASTX (NCBI) was used to study the homology of the nucleotide sequence
and the deduced amino acid sequence. DNAMAN5.2.2 was used to conduct
multiple sequence alignment. The ORF finder (NCBI) was used to search for an
Table 1. Primers used to clone and expression analysis of RrMYB6 in R. rugosa.
Primer sequence (5’ to 3’)
Purpose
Annealing
temperatures/˚C
Cloning of the
Middle Fragment
3’RACE PCR
ORF PCR
Real-time
Quantitative PCR
63.6
57.2
62.2
70.4
63.1
54.8
64.0
61.3
56.1
57.9
Name
6-F
6-R
6-3’-F
B26
RrMYB6-F
RrMYB6-R
Actin-F
Actin-R
AAGCTCACACCAACAAAGGGG
TCAAGGGTCTGTGAAATCTGT
TGTCAGTGCACAAATGGCTTC
GACTCGAGTCGACATCGATTTTTTTT
TTTTTTTTT
ATGGGAAGGTCACCTTGCTGT
CTATGAATTCAAGGGTCTGTG
CACTTAGCACCTTCCAGCAGATGT
CTACAACAGCAGACCTGAGTTCACT
RrMYB6-Q-F
CACCGAAGAAGAAGACGAG
RrMYB6-Q-R
TTCATTGTCGGTTCGTCC
DOI: 10.4236/as.2018.93026
376
Agricultural Sciences
�
K. Zou et al.
open reading frame, and the Conserved Domains database (NCBI) was used to
analyze the conserved domains. ExPaSy-SOPMA was used to predict protein
secondary structure. The ProtParam Tool was used to analyze protein physical
and chemical properties. Furthermore, the ProtScale was used to predict hydro-
philic or hydrophobic protein proper-ties. The NetPhos 3.1 Server was used to
predict potential protein phosphorylation sites, and the NetOGlyc 4.0 Server was
used to predict potential protein glycosylation sites. In order to study the evolu-
tionary relationship between RrMYB6 and MYB6 proteins in other species, we
chose 13 MYB6 genes in different spices with high level of homology to RrMYB6
by BLAST, and constructed evolutionary tree with them by MEGA5.0 software.
The phylogenetic tree was constructed according to the neighbor-joining me-
thod, and tested by bootstrap, which was repeated 1000 times.
2.2.3. Expression Analysis of RrMYB6 in Different Tissues and Different
Cultivars
Using CFX96TM Real-Time System RT-qPCR instrument and SYBR®Premix Ex
TaqTM kit as template for real-time fluorescence quantitative analysis. The pri-
mers and internal reference primers (Actinin) are shown in Table 1. The reac-
tion system is: Mix10ul, cDNA 1 ul, 0.4 ul each of upstream and downstream
primer, adding sterilizing ddH2O up to 20 ul. The reaction procedure was: pre-
denaturation at 95˚C for 30 s; 95˚C for 30 s, 60˚C for 30 s for a total of 39 cycles.
The cycle threshold (Ct) value for each PCR reaction was calculated. After com-
pletion of the amplification steps, the melting curve was determined for each
analysis. Gene transcripts were quantified using the comparative Ct method,
which compares the transcript level of the target gene with that of the reference
gene. The gene was set up with three repeats and the experimental data were
processed by 2−ΔΔCT. And the data analysis was graphed through software Prism
5.
3. Results and Analysis
3.1. Cloning and Sequence Analysis of RrMYB6
One R2R3-MYB transcription factor, RrMYB6, was cloned from the petals of
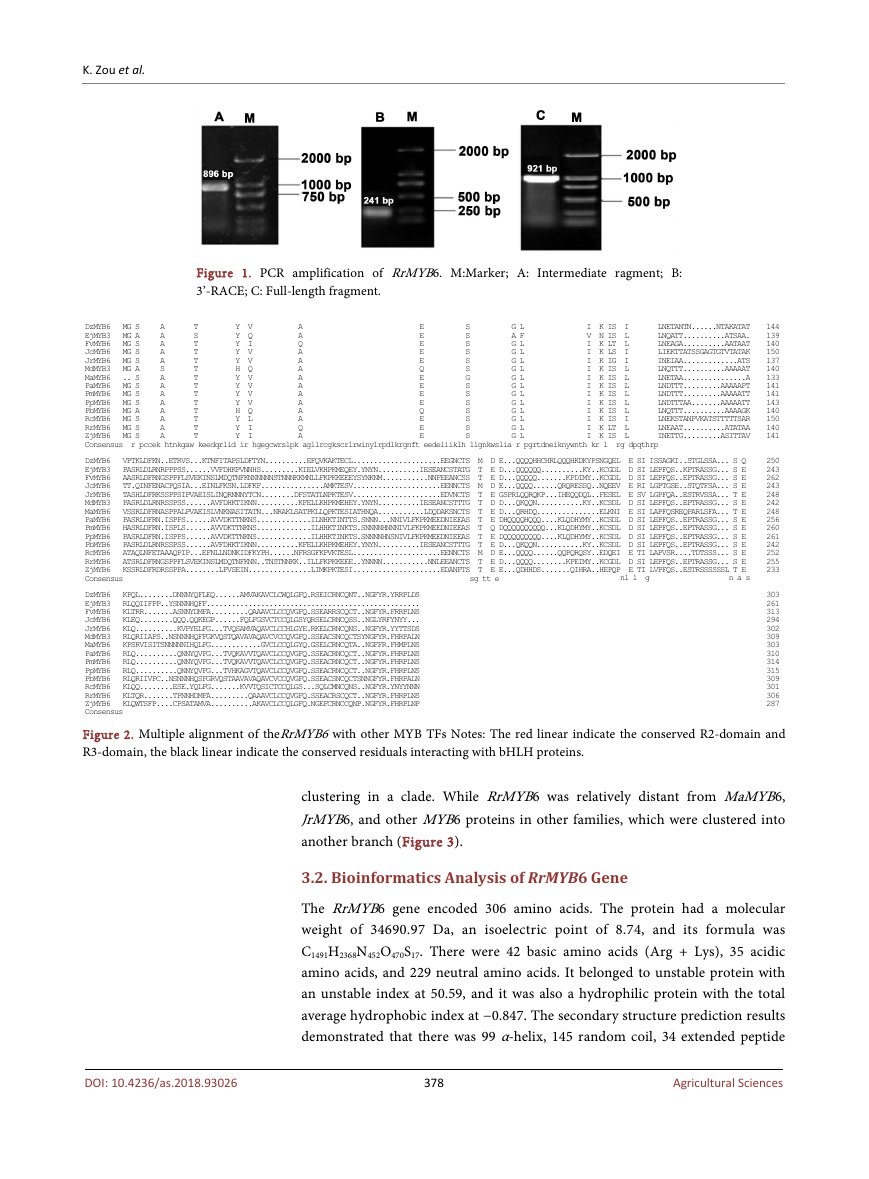
Rosa rugosa “Zi zhi”. Its Genbank accession Nos: MG745778. The RrMYB6 gene
has a full length of 1107 bp, an open reading frames (ORFs) with length of 921
bp (Figure 1), and encoding 306 amino acids.
Multiple sequence alignment analysis showed that the amino acid sequence
encoded by RrMYB6 had five conserved regions, including R2R3-MYB domain,
C 1 and C 2 suppressor motifs domain, PCCEK (N1) motif and the domain in-
teracting with bHLH protein. It is inferred that RrMYB6 is a typical R2R3-MYB
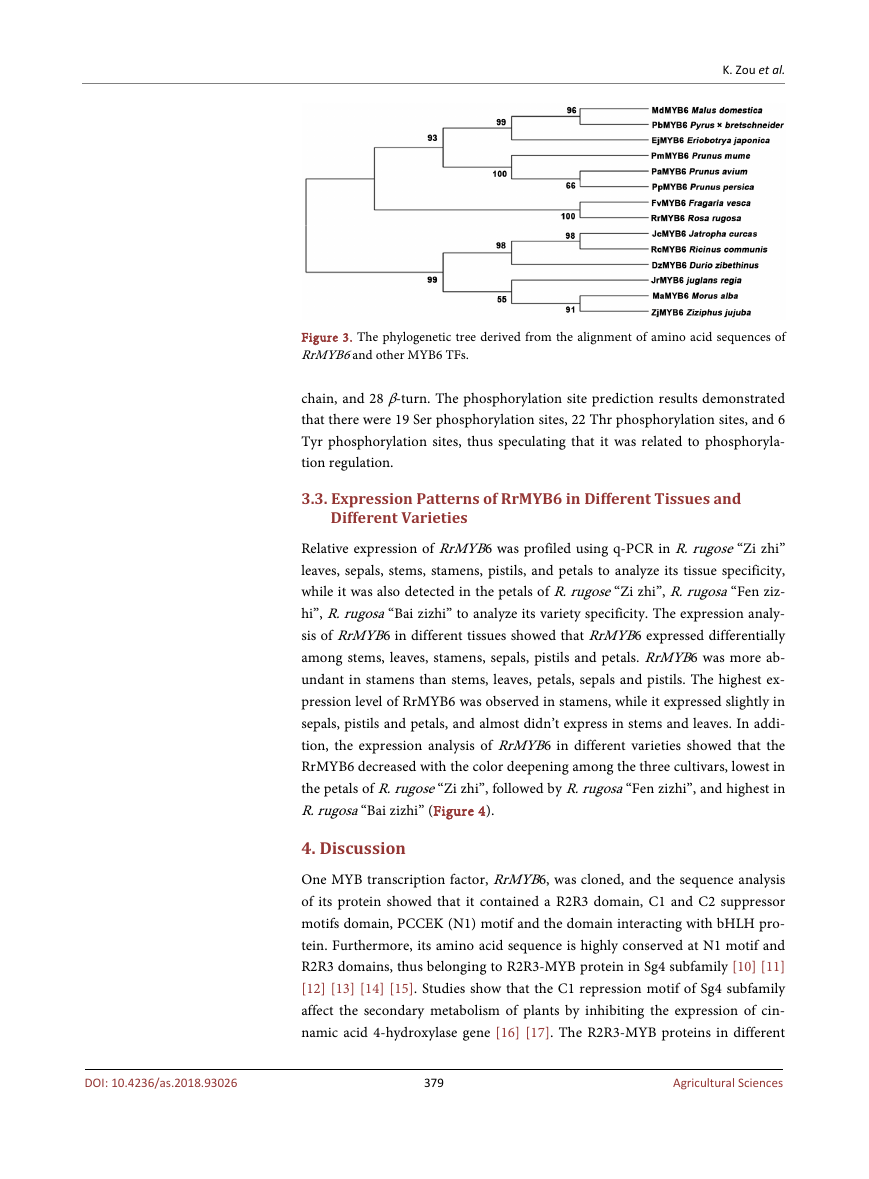
transcription factor (Figure 2). The evolutionary tree results revealed that
RrMYB6 exhibited the highest homology to FvMYB6 in Fragaria vesca. And six
MYB6 proteins from other plants in Rosaceae family, such as MdMYB6,
PmMYB6 and so on, were shown to be closely related with RrMYB6, and
377
Agricultural Sciences
DOI: 10.4236/as.2018.93026
�
K. Zou et al.
Figure 1. PCR amplification of RrMYB6. M:Marker; A: Intermediate ragment; B:
3’-RACE; C: Full-length fragment.
144
139
140
150
137
140
133
141
141
143
140
150
140
141
T
A
T
A
K
A
T
N
.
.
.
.
.
.
N
T
N
A
T
E
N
L
P
R
H
T
Q
P
D
I
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
DzMYB6
.
A
A
S
T
A
.
.
.
.
.
.
.
.
.
.
T
T
A
Q
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
N
R
K
V
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
F
R
A
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
Q
R
I
Y
D
I
L
R
Q
D
E
E
K
S
W
A
G
K
N
T
H
A
K
E
C
C
P
A
R
G
M
EjMYB3
T
A
A
T
A
A
.
.
.
.
.
.
.
.
.
.
A
G
A
E
N
L
P
R
H
T
Q
P
D
L
G
R
T
L
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
Q
K
P
L
S
R
W
C
G
E
G
H
I
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
FvMYB6
K
A
T
A
T
V
T
G
T
G
A
G
S
S
T
A
T
T
K
E
I
L
P
R
H
T
Q
P
D
I
G
R
S
L
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
JcMYB6
S
T
A
.
.
.
.
.
.
.
.
.
.
.
.
.
A
A
I
E
N
I
P
R
H
T
Q
P
D
I
G
R
G
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
JrMYB6
T
A
A
A
A
A
.
.
.
.
.
.
.
.
.
.
T
T
T
Q
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
Q
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
Q
R
I
H
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
S
K
E
C
C
P
A
R
G
M
MdMYB3
A
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
A
A
T
E
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
G
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
.
.
MaMYB6
T
P
A
A
A
A
A
.
.
.
.
.
.
.
.
.
T
T
T
D
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
PaMYB6
T
T
A
A
A
A
A
.
.
.
.
.
.
.
.
.
T
T
T
D
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
PmMYB6
T
T
A
A
A
A
A
.
.
.
.
.
.
.
A
A
T
T
T
D
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
V
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
PpMYB6
K
G
A
A
A
A
.
.
.
.
.
.
.
.
.
.
T
T
T
Q
N
L
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
Q
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
Q
R
I
H
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
A
R
G
M
PbMYB6
R
A
S
T
T
T
T
T
S
T
A
K
V
P
N
A
T
S
K
E
N
L
P
R
H
T
Q
P
D
I
G
R
S
I
L
K
R
K
H
I
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
L
R
I
Y
D
I
R
L
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
RcMYB6
A
A
T
A
T
A
.
.
.
.
.
.
.
.
.
.
T
A
A
E
N
L
P
R
H
T
Q
P
D
L
G
R
T
L
L
K
R
K
H
I
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
Q
K
P
L
S
R
W
C
G
E
G
H
I
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
RrMYB6
V
A
T
T
I
S
A
.
.
.
.
.
.
.
.
.
G
T
T
E
N
I
P
R
H
T
Q
P
D
L
G
R
S
I
L
K
R
K
I
H
T
N
W
Y
N
K
I
E
N
D
T
R
G
P
L
R
G
A
I
L
S
W
K
N
G
L
L
S
H
L
K
I
I
L
E
D
E
E
E
T
F
N
G
R
K
L
D
P
R
L
Y
N
I
W
R
L
R
C
S
K
G
C
R
L
L
G
A
A
K
P
L
S
R
W
C
G
E
G
H
I
R
I
Y
D
I
L
R
Q
D
E
E
K
T
W
A
G
K
N
T
H
A
K
E
C
C
P
S
R
G
M
ZjMYB6
p
r
h
t
q
p
d
g
r
l
r
k
h
t
n
w
y
n
k
i
e
n
d
t
r
g
p
r
a
i
l
s
w
k
n
g
l
l
h
l
k
i
i
l
e
d
e
e
t
f
n
g
r
k
l
d
p
r
l
y
n
i
w
r
l
r
c
s
k
g
c
r
l
l
g
a
k
p
l
s
r
w
c
g
e
g
h
r
i
d
i
l
r
q
d
e
e
k
w
a
g
k
n
t
h
k
e
c
c
p
r
Consensus
S
Q
A
S
N
.
.
.
A
S
S
L
G
T
S
.
.
A
S
S
I
G
I
I
K
G
S
L
E
L
N
L
E
Q
G
N
S
P
Y
K
D
K
H
Q
Q
Q
L
R
H
C
H
H
Q
Q
Q
Q
.
.
.
E
E
D
T
T
M
G
S
S
T
C
N
G
E
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
L
C
E
T
K
A
K
V
Q
.
.
.
F
E
.
.
.
.
.
.
.
N
Y
T
F
D
L
S
P
A
T
I
F
N
T
K
.
.
.
S
V
R
T
E
.
.
N
K
F
D
L
K
T
P
V
DzMYB6
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
K
.
F
P
E
L
G
I
.
S
Q
S
L
D
L
N
L
D
G
C
K
.
.
Y
K
.
.
.
.
.
.
.
.
.
.
Q
Q
Q
Q
Q
Q
.
.
.
D
E
E
T
T
T
G
S
G
T
A
T
S
C
N
A
E
S
E
I
.
.
.
.
.
.
.
.
.
.
N
Y
N
Y
.
Y
E
Q
E
M
K
P
H
.
K
V
L
E
I
K
.
.
.
.
.
.
.
.
S
H
N
N
V
P
K
H
D
F
V
V
.
.
.
.
.
.
S
S
P
P
P
R
N
R
L
D
L
R
S
A
P
EjMYB3
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
E
P
.
.
S
Q
F
E
L
G
I
S
L
D
L
N
L
D
G
C
K
.
.
Y
M
I
D
P
K
.
.
.
.
.
.
.
Q
Q
Q
Q
Q
.
.
.
D
E
E
T
T
T
G
S
S
S
C
N
A
E
E
F
N
N
.
.
.
.
.
.
.
.
.
.
.
M
N
K
N
Y
S
Y
E
E
E
K
K
P
K
K
F
L
L
N
M
K
N
N
N
T
S
N
N
N
N
N
N
K
F
N
T
Q
D
M
L
S
N
I
K
E
V
S
L
F
P
P
S
G
N
R
F
D
L
R
S
A
A
FvMYB6
S
E
A
S
N
.
.
.
A
S
F
T
Q
T
S
P
.
.
E
S
G
T
G
L
G
I
R
L
E
L
N
V
E
E
Q
N
.
.
Q
S
S
E
R
Q
R
Q
.
.
.
.
.
.
Q
Q
Q
Q
.
.
.
E
E
D
T
T
M
G
S
S
T
C
N
N
E
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
V
S
E
T
K
M
.
A
.
.
.
.
.
.
.
.
.
.
.
.
.
.
F
K
F
D
L
.
N
S
K
F
L
N
I
E
.
.
.
A
I
S
Q
P
C
A
N
E
F
N
I
Q
.
T
T
JcMYB6
S
E
A
T
N
.
.
.
S
S
A
R
V
.
E
S
T
L
S
V
G
L
G
P
F
Q
.
E
A
L
N
L
E
S
E
F
.
.
L
Q
D
Q
Q
E
H
I
.
.
.
P
K
Q
R
Q
Q
L
R
P
S
G
E
E
T
T
T
G
S
S
T
C
N
V
D
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
S
V
K
T
E
P
.
.
.
.
.
D
F
S
T
A
T
L
N
.
.
.
C
N
T
Y
N
N
N
R
Q
N
I
L
S
I
E
A
V
P
I
S
P
P
S
S
K
R
F
D
L
H
S
A
T
JrMYB6
S
E
A
S
N
.
.
.
S
S
G
R
A
P
T
S
I
G
L
E
P
F
.
.
E
L
Q
S
D
L
L
N
D
S
C
K
.
.
Y
K
.
.
.
.
.
.
.
.
.
.
.
N
Q
Q
K
Q
.
.
.
D
E
D
T
T
T
G
S
G
T
T
T
S
C
N
A
E
S
E
I
.
.
.
.
.
.
.
.
.
.
N
Y
N
Y
.
Y
E
K
M
E
H
P
.
L
L
K
H
.
.
.
.
.
.
K
F
E
.
.
.
N
N
K
I
T
K
H
D
F
V
A
.
.
.
.
.
.
S
S
P
S
S
R
N
R
L
D
L
R
S
A
P
MdMYB3
S
E
A
T
N
.
.
.
F
A
R
L
S
P
A
S
I
G
L
A
P
R
E
Q
E
L
F
Q
S
L
N
I
N
K
L
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Q
D
H
R
Q
.
.
.
D
E
E
T
T
T
G
S
S
T
C
N
S
K
A
D
Q
D
L
.
.
.
.
.
.
.
.
.
.
.
A
Q
N
H
T
A
I
T
E
S
P
K
L
S
K
L
L
Q
.
N
R
A
K
A
T
P
.
.
N
T
A
T
I
S
A
N
K
N
V
L
S
I
E
A
V
P
L
A
P
P
S
A
N
R
F
D
L
R
S
S
V
MaMYB6
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
E
.
.
P
E
L
G
I
S
S
Q
F
L
D
L
N
L
D
S
C
K
.
.
Y
M
Y
H
D
Q
L
K
.
.
.
.
Q
Q
Q
H
Q
Q
Q
Q
H
D
E
E
T
T
T
G
S
S
A
E
E
I
N
D
E
E
M
K
P
K
F
L
V
I
N
N
.
.
.
N
N
N
S
.
S
T
T
N
I
T
K
H
N
L
I
.
.
.
.
.
.
.
.
.
.
.
.
.
S
N
K
N
T
T
K
D
V
V
A
.
.
.
.
.
.
S
P
P
S
I
.
N
R
F
D
L
R
S
A
P
PaMYB6
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
E
.
.
P
E
L
G
I
S
S
Q
F
L
D
L
N
L
D
S
C
K
.
.
Y
M
Y
H
D
Q
L
K
.
.
.
Q
Q
Q
Q
Q
Q
Q
Q
Q
Q
D
E
Q
T
T
T
G
S
S
A
E
E
I
N
D
E
E
M
K
P
K
F
L
V
I
N
N
N
H
N
N
N
N
S
.
S
T
K
N
I
T
K
H
H
L
I
.
.
.
.
.
.
.
.
.
.
.
.
.
S
N
K
N
T
T
K
D
V
V
A
.
.
.
.
.
.
S
L
P
S
I
.
N
R
F
D
L
R
S
A
H
PmMYB6
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
E
.
.
S
Q
F
P
E
L
G
I
S
L
D
L
N
L
D
S
C
K
.
.
Y
M
Y
H
D
Q
L
K
.
.
.
.
Q
Q
Q
Q
Q
Q
Q
Q
Q
D
E
E
T
T
T
G
S
S
A
E
E
I
N
D
E
E
M
K
P
K
F
L
V
I
N
S
N
H
N
N
N
N
S
.
S
T
K
N
I
T
K
H
H
L
I
.
.
.
.
.
.
.
.
.
.
.
.
.
S
N
K
N
T
T
K
D
V
V
A
.
.
.
.
.
.
S
P
P
S
I
.
N
R
F
D
L
R
S
A
P
PpMYB6
S
E
A
S
N
.
.
.
G
S
S
A
R
T
P
E
.
.
S
F
P
E
L
I
S
L
G
Q
D
L
N
L
D
S
C
K
.
.
Y
K
.
.
.
.
.
.
.
.
.
.
.
N
Q
Q
K
Q
.
.
.
D
E
E
T
T
T
G
S
G
T
T
T
S
C
N
A
E
S
E
I
.
.
.
.
.
.
.
.
.
.
N
Y
N
Y
.
Y
E
H
E
M
K
P
H
K
L
L
E
K
.
.
.
.
.
.
.
F
.
.
.
N
N
K
I
T
K
H
D
F
V
A
.
.
.
.
.
.
S
S
P
S
S
R
N
R
L
D
L
R
S
A
P
PbMYB6
S
E
A
S
N
.
.
.
S
S
S
T
D
T
.
.
.
.
P
A
L
I
T
L
G
R
S
V
E
L
N
I
E
Q
D
E
.
.
Y
S
Q
R
Q
P
Q
Q
.
.
.
.
.
.
Q
Q
Q
Q
.
.
.
E
E
D
T
T
M
G
S
S
T
C
N
N
E
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
L
S
E
T
K
V
P
K
F
G
S
N
.
.
.
.
.
R
F
.
H
P
Y
K
F
D
I
K
N
D
N
L
L
N
F
E
.
.
.
P
I
P
Q
A
A
A
T
E
F
N
L
Q
A
T
A
RcMYB6
S
E
A
S
N
.
.
.
S
S
G
T
R
A
P
I
L
S
G
L
E
P
F
S
.
.
E
D
Q
L
N
L
D
G
C
K
.
.
Y
M
I
E
P
K
.
.
.
.
.
.
.
.
Q
Q
Q
Q
.
.
.
D
E
E
T
T
T
G
S
S
T
C
N
A
E
E
L
N
N
.
.
.
.
.
.
.
.
.
.
.
N
N
N
N
Y
.
.
E
E
K
K
E
K
P
S
T
N
N
K
K
L
L
F
T
N
.
.
I
.
.
N
N
K
F
N
T
Q
D
M
L
S
N
I
K
E
V
S
L
F
P
P
S
G
N
R
F
D
L
R
S
T
A
RrMYB6
S
E
A
T
N
L
S
S
S
S
S
T
R
S
S
T
I
G
L
V
P
.
.
E
E
L
F
Q
S
L
N
Q
P
P
E
H
.
.
A
R
H
I
Q
.
.
.
.
.
.
.
S
D
H
H
D
Q
.
.
.
E
E
E
T
T
T
G
S
S
T
F
N
A
D
E
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
I
K
T
E
S
K
P
.
.
.
L
I
M
.
.
.
.
.
.
.
.
.
.
.
.
N
I
E
S
V
P
L
.
.
.
.
.
.
.
.
A
P
P
S
S
R
D
R
F
D
L
R
S
S
K
ZjMYB6
s
a
n
g
l
l
n
e
t
t
g
s
Consensus
S
D
L
P
R
R
Y
.
R
Y
F
G
N
.
.
T
N
Q
C
N
R
C
I
E
S
R
.
Q
F
G
L
Q
W
C
L
C
V
A
K
A
V
M
A
.
.
.
.
.
.
Q
E
L
F
Q
Y
N
N
N
D
.
.
.
.
.
.
.
.
L
Q
P
K
DzMYB6
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
F
F
Q
H
N
N
N
S
Y
.
.
P
P
F
I
I
Q
Q
L
R
EjMYB3
S
N
L
P
R
R
F
.
R
Y
F
G
N
.
.
T
C
Q
C
S
R
R
A
E
S
S
.
Q
F
G
V
Q
C
C
L
C
V
A
A
A
Q
.
.
.
.
.
.
.
.
.
A
F
M
D
Y
N
N
S
A
.
.
.
.
.
.
.
R
R
T
L
K
FvMYB6
.
.
.
Y
Y
N
Y
F
R
Y
L
G
N
.
.
S
S
Q
C
N
R
C
L
E
S
R
Q
Y
S
G
L
Q
C
C
T
C
V
S
G
F
L
Q
F
.
.
.
.
.
.
P
G
E
K
Q
Q
.
Q
Q
Q
.
.
.
.
.
.
.
.
Q
E
L
K
JcMYB6
S
D
S
T
T
Y
Y
.
R
Y
F
G
N
.
.
S
N
Q
C
N
R
C
L
E
K
R
.
E
Y
G
L
H
C
C
L
C
V
A
Q
A
V
M
A
S
Q
V
T
.
.
.
G
F
L
E
Y
P
V
K
.
.
.
.
.
.
.
.
.
.
Q
L
K
JrMYB6
N
L
A
P
R
H
F
.
R
Y
F
G
N
Y
S
T
C
Q
C
N
S
C
A
S
E
.
S
Q
G
F
C
C
Q
V
V
C
V
A
Q
A
A
V
V
A
Q
T
S
Q
V
K
G
F
F
Q
H
N
N
N
S
N
.
.
S
P
A
I
I
R
Q
L
R
MdMYB3
S
N
L
P
M
H
F
.
R
F
F
G
N
.
.
A
T
Q
C
N
R
C
L
S
E
G
Q
.
G
Y
C
Q
L
L
C
C
V
.
G
.
.
.
.
.
.
.
.
.
.
.
G
F
L
Q
H
I
N
N
N
N
N
S
T
I
S
I
V
R
S
P
K
MaMYB6
S
N
L
P
R
H
F
.
R
Y
F
G
N
.
.
T
C
Q
N
C
R
C
A
S
E
S
.
G
F
Q
C
Q
V
L
C
C
V
Q
A
V
V
T
A
K
Q
V
T
.
.
.
G
F
V
Q
Y
N
N
Q
.
.
.
.
.
.
.
.
.
.
Q
L
R
PaMYB6
S
N
L
P
R
H
F
.
R
Y
F
G
N
.
.
T
C
Q
C
N
R
C
A
E
S
S
.
Q
F
G
V
Q
C
C
L
C
V
A
Q
T
V
V
A
K
Q
V
T
.
.
.
G
F
V
Q
Y
N
N
Q
.
.
.
.
.
.
.
.
.
.
Q
L
R
PmMYB6
S
N
L
P
R
H
F
.
R
Y
F
G
N
.
.
T
C
Q
C
N
R
C
A
E
S
S
.
Q
F
G
V
Q
C
C
L
C
V
A
Q
T
V
G
A
K
H
V
T
.
.
.
G
F
V
Q
Y
N
N
Q
.
.
.
.
.
.
.
.
.
.
Q
L
R
PpMYB6
N
L
A
P
R
H
F
.
R
Y
F
G
N
N
S
T
C
Q
C
N
S
C
A
E
S
S
.
Q
F
G
V
Q
C
C
V
C
V
A
Q
A
V
A
V
A
A
T
S
Q
V
R
G
F
S
Q
H
N
N
N
S
N
.
.
C
P
V
I
I
R
Q
L
R
PbMYB6
N
N
N
Y
Y
N
Y
.
R
Y
F
G
N
.
.
S
N
Q
C
N
M
C
L
Q
S
.
.
.
S
G
L
Q
C
C
T
C
I
S
Q
T
V
V
K
.
.
.
.
.
.
.
G
F
L
Q
Y
.
E
S
E
.
.
.
.
.
.
.
.
Q
Q
L
K
RcMYB6
S
N
L
P
R
H
F
.
R
Y
F
G
N
.
.
T
C
Q
C
S
R
C
E
A
S
S
.
F
Q
V
G
C
C
Q
C
L
V
A
.
Q
A
A
.
.
.
.
.
.
.
.
A
F
M
D
H
N
N
P
T
.
.
.
.
.
.
.
R
Q
T
L
K
RrMYB6
P
N
L
P
R
H
F
.
R
Y
F
G
N
.
P
N
Q
C
C
N
R
C
E
F
G
.
N
F
Q
L
G
C
C
Q
C
L
V
A
.
A
K
.
.
.
.
.
.
.
.
.
A
V
M
A
T
A
S
P
C
.
.
.
.
P
F
S
T
W
Q
L
K
ZjMYB6
Consensus
Figure 2. Multiple alignment of theRrMYB6 with other MYB TFs Notes: The red linear indicate the conserved R2-domain and
R3-domain, the black linear indicate the conserved residuals interacting with bHLH proteins.
250
243
262
243
248
242
248
256
260
261
242
252
255
233
303
261
313
294
302
309
303
310
314
315
309
301
306
287
clustering in a clade. While RrMYB6 was relatively distant from MaMYB6,
JrMYB6, and other MYB6 proteins in other families, which were clustered into
another branch (Figure 3).
3.2. Bioinformatics Analysis of RrMYB6 Gene
The RrMYB6 gene encoded 306 amino acids. The protein had a molecular
weight of 34690.97 Da, an isoelectric point of 8.74, and its formula was
C1491H2368N452O470S17. There were 42 basic amino acids (Arg + Lys), 35 acidic
amino acids, and 229 neutral amino acids. It belonged to unstable protein with
an unstable index at 50.59, and it was also a hydrophilic protein with the total
average hydrophobic index at −0.847. The secondary structure prediction results
demonstrated that there was 99 α-helix, 145 random coil, 34 extended peptide
378
Agricultural Sciences
DOI: 10.4236/as.2018.93026
�
K. Zou et al.
Figure 3. The phylogenetic tree derived from the alignment of amino acid sequences of
RrMYB6 and other MYB6 TFs.
chain, and 28 β-turn. The phosphorylation site prediction results demonstrated
that there were 19 Ser phosphorylation sites, 22 Thr phosphorylation sites, and 6
Tyr phosphorylation sites, thus speculating that it was related to phosphoryla-
tion regulation.
3.3. Expression Patterns of RrMYB6 in Different Tissues and
Different Varieties
Relative expression of RrMYB6 was profiled using q-PCR in R. rugose “Zi zhi”
leaves, sepals, stems, stamens, pistils, and petals to analyze its tissue specificity,
while it was also detected in the petals of R. rugose “Zi zhi”, R. rugosa “Fen ziz-
hi”, R. rugosa “Bai zizhi” to analyze its variety specificity. The expression analy-
sis of RrMYB6 in different tissues showed that RrMYB6 expressed differentially
among stems, leaves, stamens, sepals, pistils and petals. RrMYB6 was more ab-
undant in stamens than stems, leaves, petals, sepals and pistils. The highest ex-
pression level of RrMYB6 was observed in stamens, while it expressed slightly in
sepals, pistils and petals, and almost didn’t express in stems and leaves. In addi-
tion, the expression analysis of RrMYB6 in different varieties showed that the
RrMYB6 decreased with the color deepening among the three cultivars, lowest in
the petals of R. rugose “Zi zhi”, followed by R. rugosa “Fen zizhi”, and highest in
R. rugosa “Bai zizhi” (Figure 4).
4. Discussion
One MYB transcription factor, RrMYB6, was cloned, and the sequence analysis
of its protein showed that it contained a R2R3 domain, C1 and C2 suppressor
motifs domain, PCCEK (N1) motif and the domain interacting with bHLH pro-
tein. Furthermore, its amino acid sequence is highly conserved at N1 motif and
R2R3 domains, thus belonging to R2R3-MYB protein in Sg4 subfamily [10] [11]
[12] [13] [14] [15]. Studies show that the C1 repression motif of Sg4 subfamily
affect the secondary metabolism of plants by inhibiting the expression of cin-
namic acid 4-hydroxylase gene [16] [17]. The R2R3-MYB proteins in different
379
Agricultural Sciences
DOI: 10.4236/as.2018.93026
�
K. Zou et al.
DOI: 10.4236/as.2018.93026
(a) (b)
Figure 4. Relative expression levels of RrMYB6.
spices have different effects on anthocyanin synthesis. For example, the overex-
pression of FaMYB1 in tobacco made its flower color shallower with the antho-
cyanins decreased significantly [18], while the MdMYB3 isolated from Malus
domstica exhibited oppositely [19] [20]. Evolutionary analysis showed that
RrMYB6 shared the closet homology to FvMYB6, so it was speculated that
RrMYB6 could regulate the anthocyanin synthesis in R. rugosa. Specifically,
whether it exhibited in a positive way or negative way needs further discussion.
The bioinformatics analysis showed that in the secondary structure of
RrMYB6 protein, α-helix and random coil both accounted for considerable
proportion. However, the proportion of extended strand and β-turn was smaller.
Studies have proved that α-helix plays an important role in the R motif of the
MYB domain. Each R motif is generally composed of three α-helix, in which the
α-helix at the secondary and third position will form the HTH structure. And
then combine with the first R motif, further forming a HTH domain with a hy-
drophobic core. What’s more, the third alpha helices in R motif have a role of
identifying DNA, which makes the MYB protein more specific. Therefore, it can
be further determined that RrMYB6 gene belongs to R2R3-MYB type [21] [22].
The irregular curl structure is very beneficial to the binding of cell and water
molecule, and both belong to hydrophilic protein. It can be inferred that both of
them may protect the plant from osmotic stress. In addition, RrMYB6 contains a
certain phosphorylation site, which may be related to the regulation of phos-
phorylation.
The qRT-PCR results showed that RrMYB6 expressed slightly in the petals of
R. rugose “Zi zhi”, followed by R. rugosa “Fen zizhi”, and highly expressed in R.
rugosa “Bai zizhi”. Furthermore, RrMYB6 exhibited a characteristic low expres-
sion in petals while expressed at a high level in stamen in the different tissues of
R. rugosa “Zi zhi”. Studies show that there are positive and negative mechanisms
for anthocyanin biosynthesis regulation by MYB protein in plants [22]. The
MdMYB isolated from Malus domestica regulates the synthesis of anthocyanin.
Moreover, lightness is the essential factor in the regulation of biosynthetic gene
by MdMYB. Overexpression of MdMYB10 isolated from leaves and pulp can in-
380
Agricultural Sciences
�
K. Zou et al.
crease anthocyanin accumulation in apple seedlings. What’s more, the overex-
pression of MdMYB16, MdMYB17 and MdMYB111 in tobacco could inhibit the
activity of DFR promoter and thus inhibiting anthocyanin synthesis [23] [24]. Li
also find that MYB protein has positive and negative mechanisms for anthocya-
nin synthesis in Morus alba [25]. The expression of RrMYB6 gene decreased
with the deepening of petal color. And in the expression analysis of R. rugosa “Zi
zhi” tissues, the expression of petals was very low, which was consistent with the
analysis of expression among varieties. The gene expressed slightly in the petals,
which may caused by different flowering stages. These results suggested that the
regulation of RrMYB6 gene on anthocyanin biosynthesis in R. rugosa was a neg-
ative regulation mechanism. The regulation of genes involved in this mechanism
needs further verification.
In conclusion, one R2R3-MYB TF, RrMYB6, was isolated from R. rugosa and
was found to be involved in regulating anthocyanin biosynthetic pathway. The
results of this study provided important information on the anthocyanin synthe-
sis of R. rugosa. In future work, we will test whether the overexpression of
RrMYB6 leads to anthocyanin accumulation in Arabidopsis thaliana and Nico-
tiana tabacum.
Acknowledgements
This work was funded by Shandong Province Agricultural Engineering project
of breeding ([2014] No. 96).
References
[1] Buer, C.S., Imin, N. and Djordjevic, M.A. (2010) Flavonoids: New Roles for Old
Molecules. Journal of Integrative Plant Biology, 52, 98-111.
https://doi.org/10.1111/j.1744-7909.2010.00905.x
[2] Schwinn, K.E., Boase, M.R., Bradley, J.M., et al. (2014) MYB and bHLH transcrip-
tion Factor Transgenes Increase Anthocyanin Pigmentation in Petunia and Lisian-
thus Plants, and the Petunia Phenotypes Are Strongly Enhanced under Field Condi-
tions. Frontiers in Plant Science, 5, 603. https://doi.org/10.3389/fpls.2014.00603
Ibraheem, F., Gaffoor, I., Tan, Q., et al. (2015) A Sorghum MYB Transcription Fac-
tor Induces 3-deoxyanthocyanidins and Enhances Resistance against Leaf Blights in
Maize. Molecules, 20, 2388-2404. https://doi.org/10.3390/molecules20022388
[3]
[4] Schwinn, K., Venail, J., Shang, Y., et al. (2006) A Small Family of MYB-Regulatory
Genes Controls Floral Pigmentation Intensity and Patterning in the Genus Antirr-
hinum. The Plant Cell, 18, 831-851. https://doi.org/10.1105/tpc.105.039255
[5] Ravaglia, D., Espley, R., Henry, K.R., et al. (2013) Transcriptional Regulation of
Flavonoid Biosynthesis in Nectarine (Prunus persica) by a Set of R2R3-MYB Tran-
scription Factors. BMC Plant Biology, 13, 68.
https://doi.org/10.1186/1471-2229-13-68
[6] Li, L., Ban, Z.J., Li, X.H., et al. (2012) Differential Expression of Anthocyanin Bio-
synthetic Genes and Transcription Factor PcMYB10 in Pears (Pyrus communis L.).
Plos One, 7, e46070. https://doi.org/10.1371/journal.pone.0046070
[7] Huang, W., Sun, W., Lv, H., et al. (2013) A R2R3-MYB Transcription Factor from
381
Agricultural Sciences
DOI: 10.4236/as.2018.93026
�














 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc