A v a i l a b l e a t w w w . s c i e n c e d i r e c t . c o m
INFORMATION PROCESSING IN AGRICULTURE xxx (xxxx) xxx
j o u r n a l h o m e p a g e : w w w . e l s e v i e r . c o m / l o c a t e / i n p a
Automatic grape leaf diseases identification via
UnitedModel based on multiple convolutional
neural networks
Miaomiao Ji a, Lei Zhang b, Qiufeng Wu c,*
a College of Engineering, Northeast Agricultural University, Harbin 150030, China
b School of Medicine, University of Pittsburgh, Pittsburgh 15260, USA
c College of Science, Northeast Agricultural University, Harbin 150030, China
A R T I C L E I N F O
A B S T R A C T
Article history:
Received 17 April 2019
Received in revised form
11 October 2019
Accepted 15 October 2019
Available online xxxx
Keywords:
Grape leaf diseases
Identification
Multi-network integration method
Convolutional neural network
Deep learning
Grape diseases are main factors causing serious grapes reduction. So it is urgent to develop
an automatic identification method for grape leaf diseases. Deep learning techniques have
recently achieved impressive successes in various computer vision problems, which
inspires us to apply them to grape diseases identification task. In this paper, a united con-
volutional neural networks (CNNs) architecture based on an integrated method is pro-
posed. The proposed CNNs architecture, i.e., UnitedModel is designed to distinguish
leaves with common grape diseases i.e., black rot, esca and isariopsis leaf spot from
healthy leaves. The combination of multiple CNNs enables the proposed UnitedModel to
extract complementary discriminative features. Thus the representative ability of United-
Model has been enhanced. The UnitedModel has been evaluated on the hold-out PlantVil-
lage dataset and has been compared with several state-of-the-art CNN models. The
experimental results have shown that UnitedModel achieves the best performance on var-
ious evaluation metrics. The UnitedModel achieves an average validation accuracy of
99.17% and a test accuracy of 98.57%, which can serve as a decision support tool to help
farmers identify grape diseases.
Ó 2019 China Agricultural University. Production and hosting by Elsevier B.V. on behalf of
KeAi. This is an open access article under the CC BY-NC-ND license (http://creativecommons.
org/licenses/by-nc-nd/4.0/).
1.
Introduction
Grapes, as one of the most commonly cultivated economical
fruit crops throughout the world, are widely used in the pro-
duction of wine, brandy or nonfermented drinks and are
eaten fresh or dried as raisins [1]. However, grapes are vulner-
able to various different types of diseases, such as black rot,
esca,
isariopsis leaf spot, etc. It is estimated that losses
caused by grape diseases in Georgia, USA in 2015 were
approximately $1.62 million. Around $0.5 million was spent
on diseases control and the rest was the loss caused by the
diseases [2]. Thus, early detection of grape diseases can
potentially cut losses and control costs and consequently
can improve the quality of products.
* Corresponding author.
E-mail address: qfwu@neau.edu.cn (Q. Wu).
Peer review under responsibility of China Agricultural University.
https://doi.org/10.1016/j.inpa.2019.10.003
2214-3173 Ó 2019 China Agricultural University. Production and hosting by Elsevier B.V. on behalf of KeAi.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
For decades, the diseases identification is mostly per-
formed by human. The process of recognition and diagnosis
is subjective, error-prone, costly and time-consuming.
In
addition, new diseases can occur in places where they were
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
2
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
previously unidentified and, inherently, where there is no
local expertise to combat them [3]. As a result, an automatic
identification method for grape disease identification is in
urgent demand.
The development of sophisticated instruments and fast
computational techniques have paved the way for real-time
scanning and automatic detection of anomalies in a crop
[4]. Although traditional machine learning methods have
gained some valuable experience in identification and diag-
nosis of crop diseases, they are limited to following the pipe-
lined procedures of image segmentation (such as clustering
method [5], threshold method [6], etc.), feature extraction
(such as shape, texture, color features, etc. [7]), and pattern
recognition (such as k-nearest neighbor method (KNN) [8],
support vector machine (SVM) [1], back propagation neural
network (BPNN) [9], etc.). It is difficult to select and extract
the optimal visible pathological features and thus highly
skilled engineers and experienced experts are demanded,
which is not only to a considerable extent subjective but also
leads to a great waste of manpower and financial resources.
In contrast, deep learning techniques can automatically learn
the hierarchical feature of pathologies and do not need to
manually design the feature extraction and classifier. Deep
learning method has so excellent generalization ability and
robustness that it excels in many areas: signal processing
[10], pedestrian detection [11], face recognition [12], road
crack detection [13], biomedical image analysis [14], etc. In
addition, deep learning techniques have also achieved
impressive results in the field of agriculture and benefit more
smallholders and horticultural workers, including diagnosis
of crop diseases [15], recognition of weeds [16], selection of
fine seeds [17], pest identification [18], fruit counting [19],
research on land cover [20] etc, which has contributed to deal-
ing with image classification of areas of interest. Further-
more, some applications focused on predicting future
parameters, such as crop yield [21], weather conditions [22]
and soil moisture content in the field [23]. Inspired by the
great success of CNN-based methods in image classification,
we propose an integrated model, denoted UnitedModel, for
automatic grape leaf disease identification.
The rest of this paper is organized as follows. Section 2
starts with an overview of related works. Section 3 introduces
dataset as well as data preprocessing and covers details of the
proposed UnitedModel. In Section 4, we conduct all the exper-
iments, discuss the limitation of the proposed UnitedModel
and prospect the future work. In Section 5, we conclude this
paper.
2.
Related works
Machine learning techniques were applied widely in plant
disease classification in the early stage. Li et al. [24] proposed
a method based on K-means clustering segmentation of the
grape disease images and a SVM classifier was designed
based on thirty-one effective selected features to identify
grape downy mildew disease and grape powdery mildew dis-
ease with testing recognition rates of 90% and 93.33%, respec-
tively. Significant progress has been made in the use of image
processing approaches to detect various diseases in crops.
Athanikar and Badar [9] applied Neural Network to categorize
the potato leaf image as either healthy or diseased. Their
results showed that BPNN could effectively detect the disease
spots and could classify the particular disease type with an
accuracy of 92%. The deployment of deep CNNs has especially
led to a breakthrough in plant disease classification, which
can find very high variance of pathological symptoms in
visual appearance, even high intra-class dissimilarity and
low inter-class similarity that may be only noticed by the
botanists [25]. Lee et al. [26] proposed a CNN approach to iden-
tify leaf images and reported an average accuracy of 99. 7% on
a dataset covering 44 species, but the scale of datasets was
very small. Zhang et al. [27] used GoogLeNet to address the
detection of cherry leaf powdery mildew disease and obtained
an accuracy of 99.6%. Their results also demonstrated trans-
fer learning can boost the performances of deep learning
model
[15]
fine-tuned deep learning models pre-trained on ImageNet to
identify 14 crop species and 26 diseases. The models were
evaluated on a publicly available dataset including 54,306
images of diseased and healthy plant leaves collected under
controlled conditions. They achieved the best accuracy of
99.35% on a hold-out test dataset.
in crop disease identification. Mohanty et al.
Although the basic CNN frameworks, such as AlexNet [28],
VGGNet [29], GoogLeNet [30], DenseNet [31] and ResNet [32]
have been demonstrated effective and widely used in crop
diseases classification, most previous works had troubling
boosting up the classification accuracy rate to some extent.
As a matter of fact, a single model can’t meet the further
requests in terms of precision. In large machine learning
competitions, the best results were usually achieved by the
integration of multiple models rather than by a single model.
For instance, the well-known Inception-ResNet-v2 [33] was
born out of the fusion of two excellent deep CNNs, as the
name suggests. Inspired by network in network concept
[34], we think that integration is the most straightforward
and effective way when the basic models are significantly dif-
ferent. We propose the UnitedModel in this study. The United-
Model is a integration of the powerful and popular deep
learning architectures of GoogLeNet and ResNet. We also take
advantage of transfer learning to boost accuracy as well as
reduce the training time.
3.
Materials and methods
3.1.
Dataset and preprocessing
Our dataset comes from an open access repository named
PlantVillage which focuses on plant health [35]. The dataset
used for evaluating the proposed method is composed of
healthy (171 images) and symptom images including black
rot (pathogen: Guignardia bidwellii, 476 images), esca (patho-
gen: Phaeomoniella spp, 552 images) and isariopsis leaf spot
(pathogen: Pseudocercospor a-vitis, 420 images) (see Table 1).
The identification of grape diseases is based on its leaf, not
flower, fruit or stem. For one thing, the flower and fruit of
grape only appear in a limited time while the leaf presents
most of the year. For another thing, the stem of grape can
hardly present the symptoms of the diseases timely, while
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
3
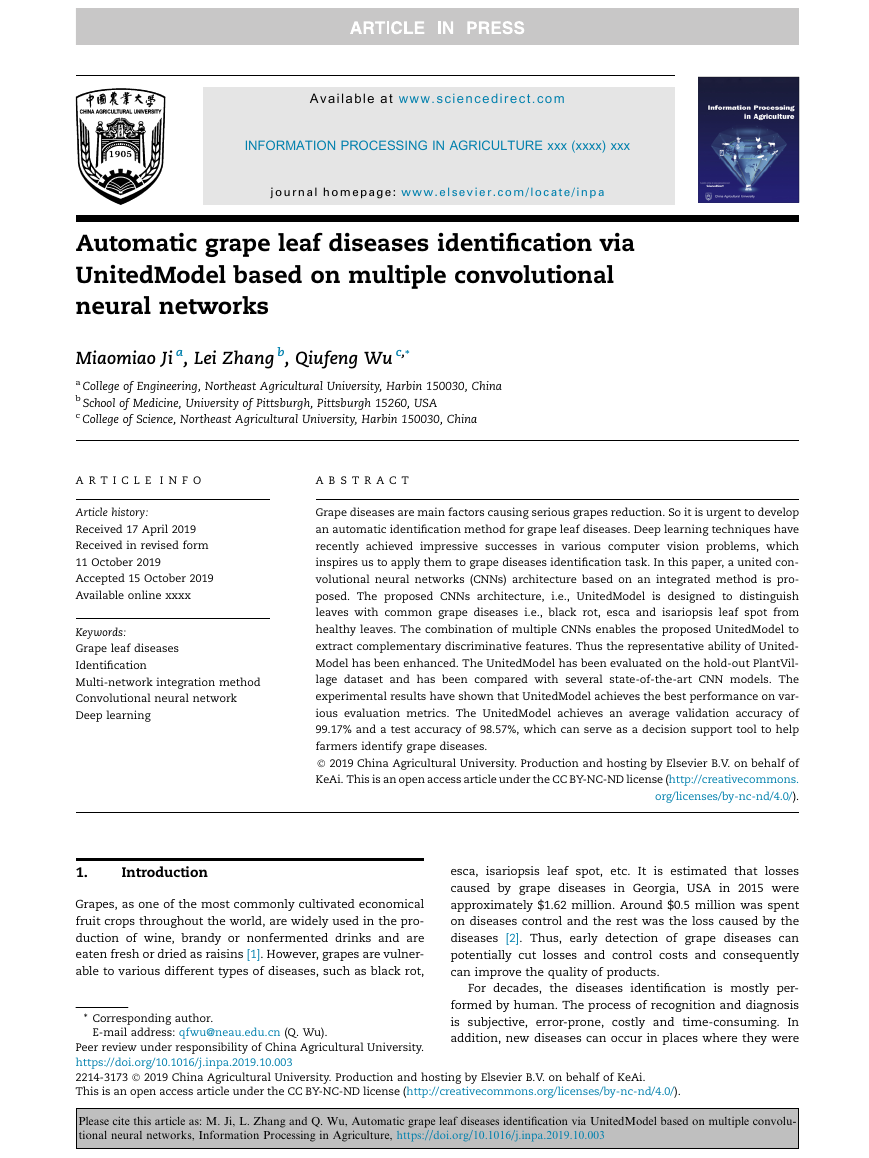
Table 1 – Introduction of grape leaf dataset.
Label Category Number Data
No data augmentation
Leaf symptoms
Illustration
augmentation
Training
samples
Validation
samples
Test
samples
1
2
3
Black rot 476
Esca
Isariopsis
leaf spot
552
420
4
Health
Total samples
171
1619
288
288
288
96
960
72
72
72
24
240
116
192
60
51
419
See Fig. 1 first row
Appear small,
brown circular lesions
Appear dark red or yellow stipes See Fig. 1 second row
Appear many small rounded,
polygonal
or irregular brown spots
–
–
See Fig. 1 fourth row
–
See Fig. 1 third row
Fig. 1 – The grape leaf images after data augmentation.
the leaf is generally sensitive to the state of plants, whose
shape, texture and color usually contains richer information.
The raw images are divided into training dataset and test
dataset. 360 Symptom and 120 healthy images are selected
for training and the rest images are used for testing. To pre-
vent over-fitting, the training dataset is further split into
training (80%) and validation data (20%). Thus the training
dataset is 960 samples in total, validation dataset is 240 sam-
ples in total and test dataset is 419 samples in total [36]. The
original images in the PlantVillage dataset are RGB images of
arbitrary size. Data preprocessing is necessary and all the
images are resized to the expected input size of the respective
networks, i.e. 224 � 224 for VGGNet, DenseNet and ResNet,
and 229 � 229 for GoogLeNet. Model optimization and predic-
tion are both performed on these rescaled images. To avoid
over-fitting and to boost the generalizability of the CNNs, data
augmentation techniques which include rotating, flipping,
shearing, zooming and colour changing are randomly per-
formed in training phase as shown in Fig. 1.
Considering that the number of healthy samples in our
work is relatively small with 171 samples in total, the propor-
tion between healthy samples and diseased samples is set to
1: 3 while the class_weight ratio of them is set to 3: 1. By this
way, the loss function can be adjusted in the training process
and healthy samples can obtain more attention.
3.2.
Identification model of grape leaf diseases
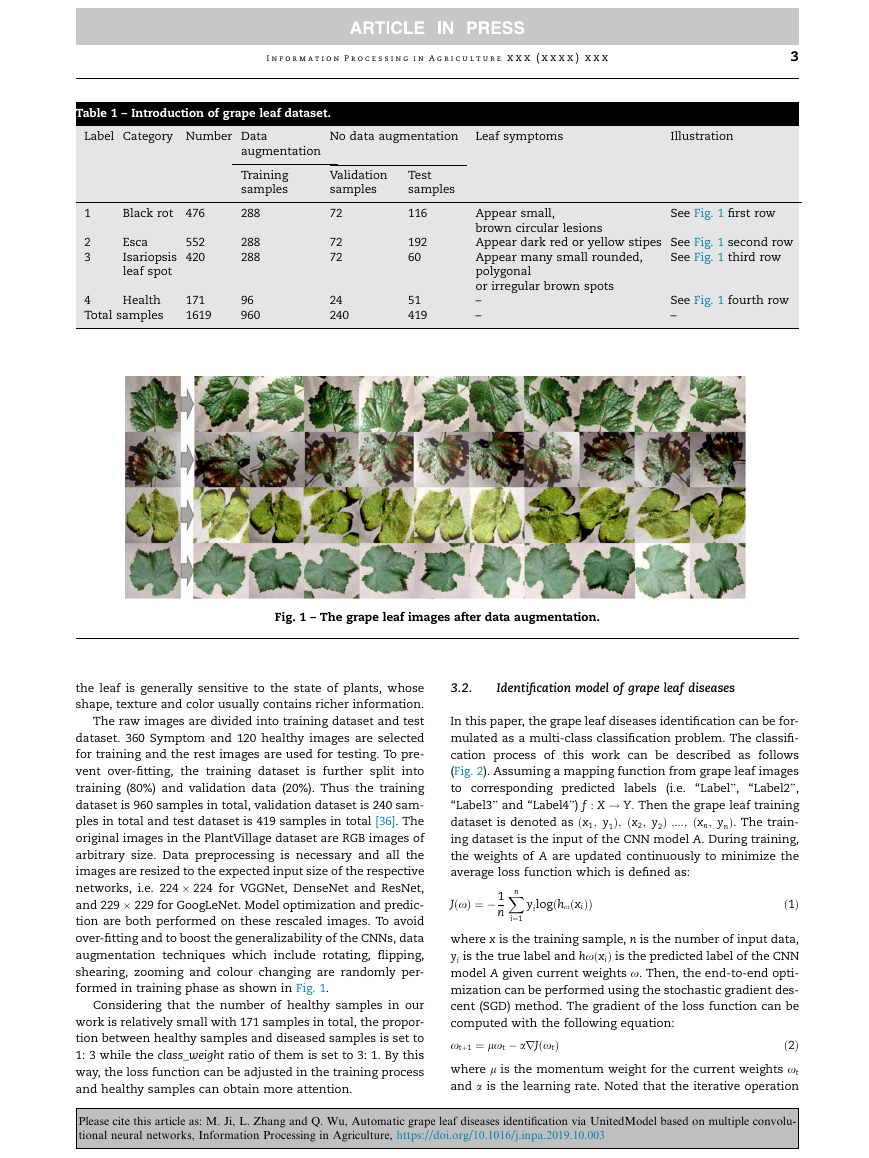
In this paper, the grape leaf diseases identification can be for-
mulated as a multi-class classification problem. The classifi-
cation process of this work can be described as follows
(Fig. 2). Assuming a mapping function from grape leaf images
‘‘Label2”,
to corresponding predicted labels (i.e.
‘‘Label3” and ‘‘Label4”) f : X ! Y: Then the grape leaf training
Þ. The train-
dataset is denoted as ðx1; y1
ing dataset is the input of the CNN model A. During training,
the weights of A are updated continuously to minimize the
average loss function which is defined as:
JðxÞ ¼ � 1
n
‘‘Label”,
Þ ;:::; ðxn; yn
yilogðhxðxiÞÞ
Þ; ðx2; y2
ð1Þ
Xn
i¼1
where x is the training sample, n is the number of input data,
yi is the true label and hxðxiÞ is the predicted label of the CNN
model A given current weights x. Then, the end-to-end opti-
mization can be performed using the stochastic gradient des-
cent (SGD) method. The gradient of the loss function can be
computed with the following equation:
xtþ1 ¼ lxt � arJðxtÞ
ð2Þ
where l is the momentum weight for the current weights xt
and a is the learning rate. Noted that the iterative operation
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
4
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
InceptionV3 is selected as one component of the UnitedMo-
del. ResNet won the first place in ILSVRC 2015 and COCO
2015 classification challenge with error rate of 3.57%, which
introduced residual units for addressing the degradation
problem of CNNs. ResNet is stacked by many residual units.
Given the size of the dataset used in this work, ResNet50
becomes our first choice to be another basic network of the
UnitedModel, which also has high performance in classifica-
tion tasks. Given sufficient data, the most straightforward
way of improving the performance of CNNs is increasing their
parameters. In this study, GoogLeNet increases its width: the
number of units at each layer while ResNet increases the
depth: the number of network layer. The integration is most
effective when the basic models are significantly different
because their combination will enable the integrated network
to capture more distinguishing feature information, thereby
improving identification accuracy.
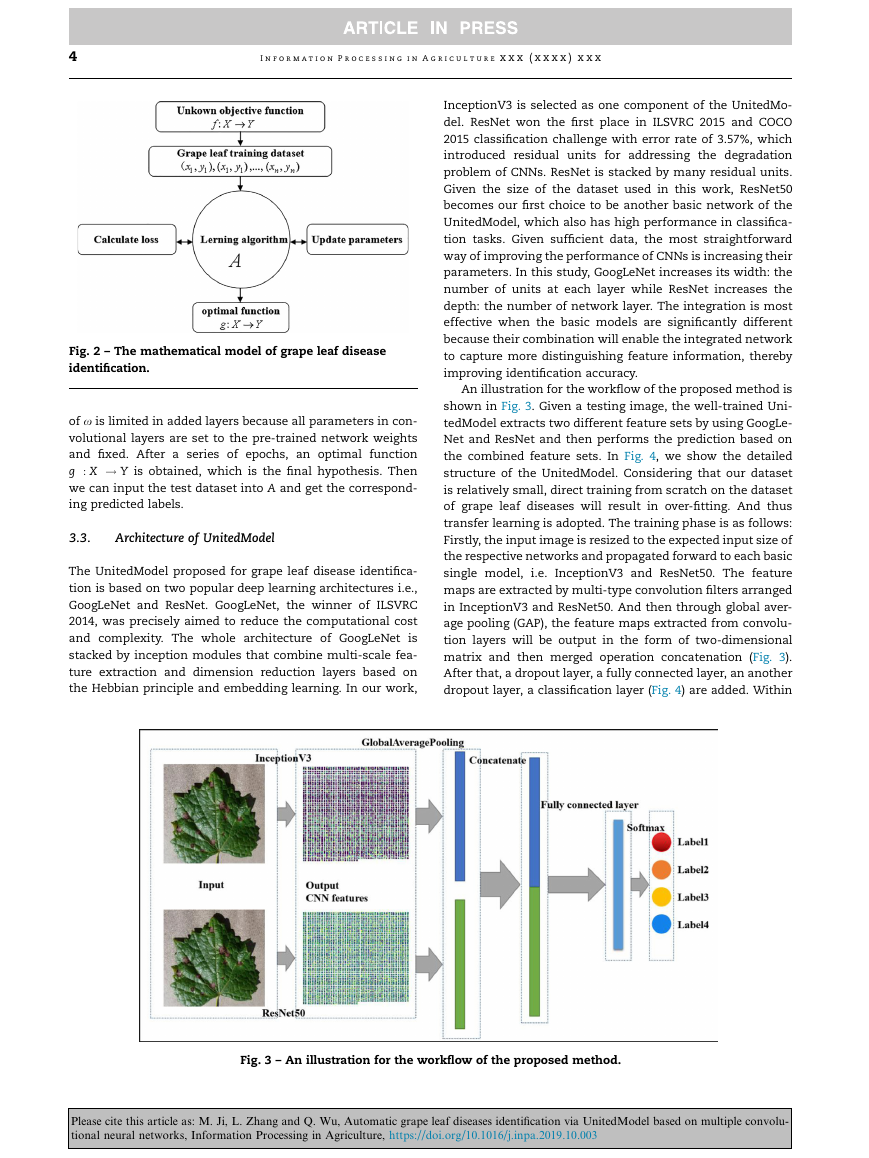
An illustration for the workflow of the proposed method is
shown in Fig. 3. Given a testing image, the well-trained Uni-
tedModel extracts two different feature sets by using GoogLe-
Net and ResNet and then performs the prediction based on
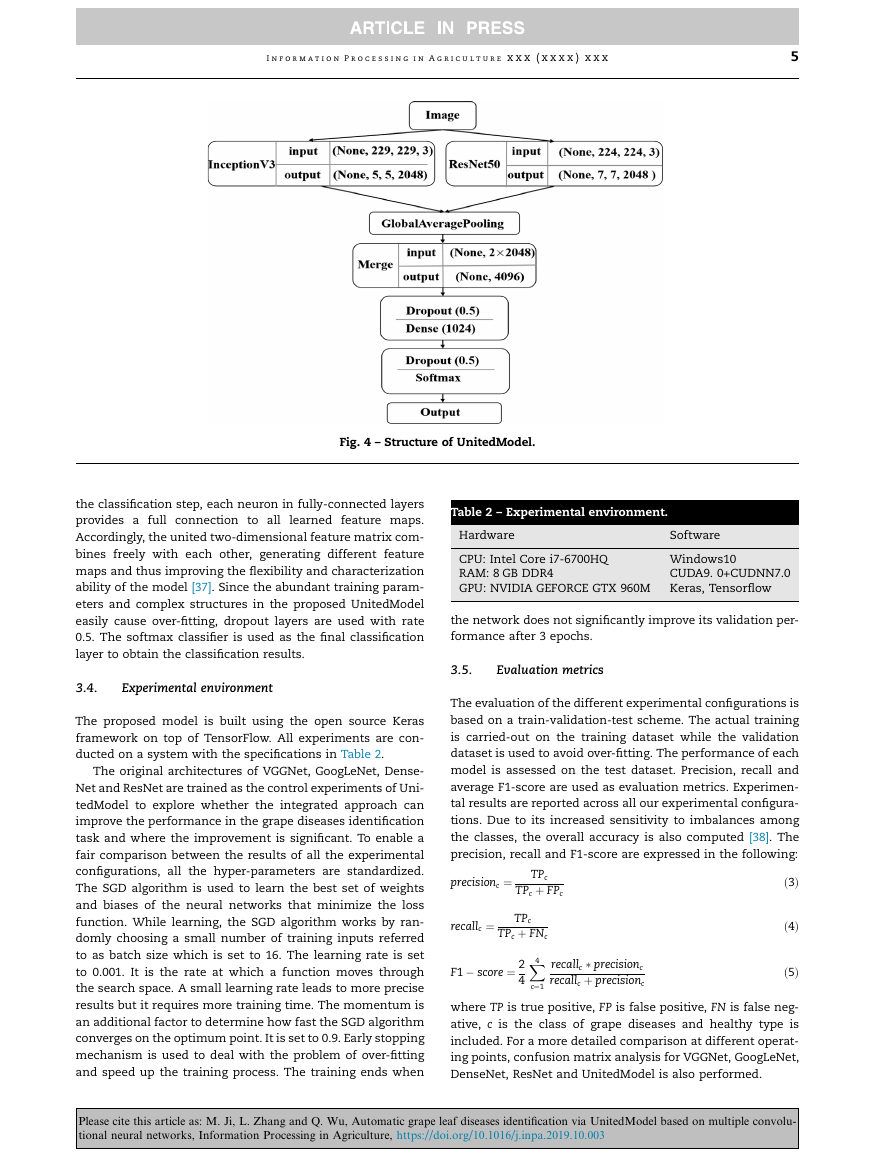
the combined feature sets. In Fig. 4, we show the detailed
structure of the UnitedModel. Considering that our dataset
is relatively small, direct training from scratch on the dataset
of grape leaf diseases will result in over-fitting. And thus
transfer learning is adopted. The training phase is as follows:
Firstly, the input image is resized to the expected input size of
the respective networks and propagated forward to each basic
single model, i.e. InceptionV3 and ResNet50. The feature
maps are extracted by multi-type convolution filters arranged
in InceptionV3 and ResNet50. And then through global aver-
age pooling (GAP), the feature maps extracted from convolu-
tion layers will be output in the form of two-dimensional
matrix and then merged operation concatenation (Fig. 3).
After that, a dropout layer, a fully connected layer, an another
dropout layer, a classification layer (Fig. 4) are added. Within
Fig. 2 – The mathematical model of grape leaf disease
identification.
of x is limited in added layers because all parameters in con-
volutional layers are set to the pre-trained network weights
and fixed. After a series of epochs, an optimal function
g : X ! Y is obtained, which is the final hypothesis. Then
we can input the test dataset into A and get the correspond-
ing predicted labels.
3.3.
Architecture of UnitedModel
The UnitedModel proposed for grape leaf disease identifica-
tion is based on two popular deep learning architectures i.e.,
GoogLeNet and ResNet. GoogLeNet, the winner of ILSVRC
2014, was precisely aimed to reduce the computational cost
and complexity. The whole architecture of GoogLeNet is
stacked by inception modules that combine multi-scale fea-
ture extraction and dimension reduction layers based on
the Hebbian principle and embedding learning. In our work,
Fig. 3 – An illustration for the workflow of the proposed method.
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
5
Fig. 4 – Structure of UnitedModel.
the classification step, each neuron in fully-connected layers
provides a full connection to all
learned feature maps.
Accordingly, the united two-dimensional feature matrix com-
bines freely with each other, generating different feature
maps and thus improving the flexibility and characterization
ability of the model [37]. Since the abundant training param-
eters and complex structures in the proposed UnitedModel
easily cause over-fitting, dropout layers are used with rate
0.5. The softmax classifier is used as the final classification
layer to obtain the classification results.
3.4.
Experimental environment
The proposed model is built using the open source Keras
framework on top of TensorFlow. All experiments are con-
ducted on a system with the specifications in Table 2.
The original architectures of VGGNet, GoogLeNet, Dense-
Net and ResNet are trained as the control experiments of Uni-
tedModel to explore whether the integrated approach can
improve the performance in the grape diseases identification
task and where the improvement is significant. To enable a
fair comparison between the results of all the experimental
configurations, all the hyper-parameters are standardized.
The SGD algorithm is used to learn the best set of weights
and biases of the neural networks that minimize the loss
function. While learning, the SGD algorithm works by ran-
domly choosing a small number of training inputs referred
to as batch size which is set to 16. The learning rate is set
to 0.001. It is the rate at which a function moves through
the search space. A small learning rate leads to more precise
results but it requires more training time. The momentum is
an additional factor to determine how fast the SGD algorithm
converges on the optimum point. It is set to 0.9. Early stopping
mechanism is used to deal with the problem of over-fitting
and speed up the training process. The training ends when
Table 2 – Experimental environment.
Hardware
Software
CPU: Intel Core i7-6700HQ
RAM: 8 GB DDR4
GPU: NVIDIA GEFORCE GTX 960M Keras, Tensorflow
Windows10
CUDA9. 0+CUDNN7.0
the network does not significantly improve its validation per-
formance after 3 epochs.
3.5.
Evaluation metrics
The evaluation of the different experimental configurations is
based on a train-validation-test scheme. The actual training
is carried-out on the training dataset while the validation
dataset is used to avoid over-fitting. The performance of each
model is assessed on the test dataset. Precision, recall and
average F1-score are used as evaluation metrics. Experimen-
tal results are reported across all our experimental configura-
tions. Due to its increased sensitivity to imbalances among
the classes, the overall accuracy is also computed [38]. The
precision, recall and F1-score are expressed in the following:
ð3Þ
precisionc ¼
TPc
TPc þ FPc
recallc ¼
F1 � score ¼ 2
4
c¼1
TPc
TPc þ FNc
X4
recallc � precisionc
recallc þ precisionc
ð4Þ
ð5Þ
where TP is true positive, FP is false positive, FN is false neg-
ative, c is the class of grape diseases and healthy type is
included. For a more detailed comparison at different operat-
ing points, confusion matrix analysis for VGGNet, GoogLeNet,
DenseNet, ResNet and UnitedModel is also performed.
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
6
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
Fig. 5 – Validation accuracy and validation loss of various architectures are compared and the number of epoch is varied in the
case of early stopping mechanism. VGGNet, GoogLeNet, DenseNet, ResNet and UnitedModel stop training with the epoch 11,
18, 11, 9 and 12, respectively.
Fig. 6 – Comparison of the average accuracy for the different architectures.
4.
Results and discussion
As illustrated in Fig. 5, UnitedModel has a tendency to consis-
tently improve in accuracy with growing number of epochs,
with no signs of performance deterioration. On the other
hand, loss curve of UnitedModel converges rapidly and has
a big gap with other single models in terms of error rate. Con-
sistent with Fig. 6, the performance of GoogLeNet, VGGNet,
DenseNet and ResNet
to
UnitedModel.
similar and all
is
inferior
As we can see in Table 3, the average precision (99.05%),
recall (98.88%) and F1-score (98.96%) of UnitedModel are the
highest among all these models. And scores of VGGNet are
the lowest with precision (94.16%), recall (93.32%) and F1-
score (94.40%), which once again proves the superiority of
integrated CNNs.
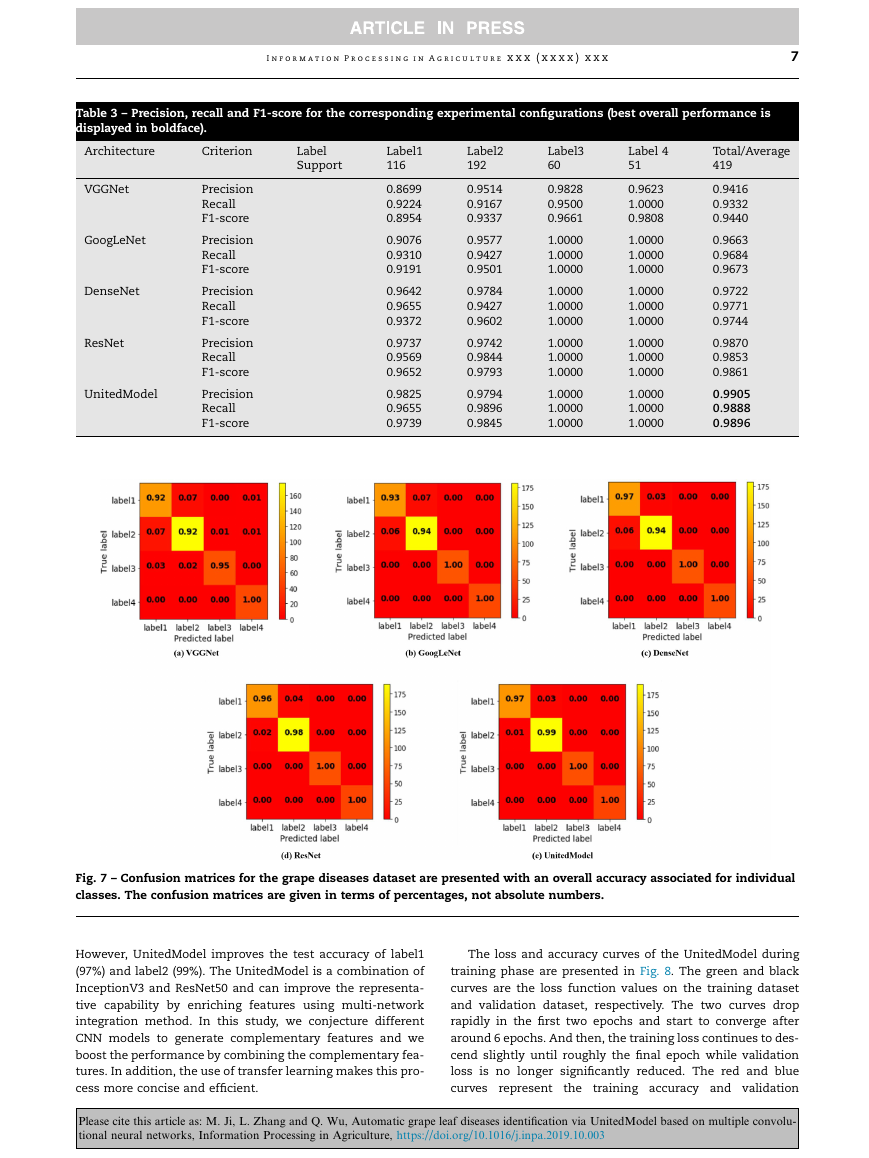
The confusion matrices of different models on test dataset
are shown in Fig. 7. The threshold is set to 0.5 and the fraction
of accurately predicted images for each class is displayed in
detail. The performance of VGGNet is poor. VGGNet tends to
distinguish diseased samples as healthy, which is extremely
harmful in actual agriculture produce. All the models except
VGGNet can distinguish label3 and label4 easily, but label1
and label2 are prone to be misclassified. The large green area
of healthy leaves and the golden appearance of leaves
infected with isariopsis leaf spot disease make them easier
to be distinguished. Meanwhile, the confusion between label1
and label2 is due to their similar pathological features.
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
7
Table 3 – Precision, recall and F1-score for the corresponding experimental configurations (best overall performance is
displayed in boldface).
Architecture
Criterion
Label
Support
VGGNet
GoogLeNet
DenseNet
ResNet
UnitedModel
Precision
Recall
F1-score
Precision
Recall
F1-score
Precision
Recall
F1-score
Precision
Recall
F1-score
Precision
Recall
F1-score
Label1
116
0.8699
0.9224
0.8954
0.9076
0.9310
0.9191
0.9642
0.9655
0.9372
0.9737
0.9569
0.9652
0.9825
0.9655
0.9739
Label2
192
0.9514
0.9167
0.9337
0.9577
0.9427
0.9501
0.9784
0.9427
0.9602
0.9742
0.9844
0.9793
0.9794
0.9896
0.9845
Label3
60
0.9828
0.9500
0.9661
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
Label 4
51
Total/Average
419
0.9623
1.0000
0.9808
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
1.0000
0.9416
0.9332
0.9440
0.9663
0.9684
0.9673
0.9722
0.9771
0.9744
0.9870
0.9853
0.9861
0.9905
0.9888
0.9896
Fig. 7 – Confusion matrices for the grape diseases dataset are presented with an overall accuracy associated for individual
classes. The confusion matrices are given in terms of percentages, not absolute numbers.
However, UnitedModel improves the test accuracy of label1
(97%) and label2 (99%). The UnitedModel is a combination of
InceptionV3 and ResNet50 and can improve the representa-
tive capability by enriching features using multi-network
integration method. In this study, we conjecture different
CNN models to generate complementary features and we
boost the performance by combining the complementary fea-
tures. In addition, the use of transfer learning makes this pro-
cess more concise and efficient.
The loss and accuracy curves of the UnitedModel during
training phase are presented in Fig. 8. The green and black
curves are the loss function values on the training dataset
and validation dataset, respectively. The two curves drop
rapidly in the first two epochs and start to converge after
around 6 epochs. And then, the training loss continues to des-
cend slightly until roughly the final epoch while validation
loss is no longer significantly reduced. The red and blue
the training accuracy and validation
curves represent
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�
8
I n f o r m a t i o n P r o c e s s i n g i n A g r i c u l t u r e x x x ( x x x x ) x x x
support tool to help farmers identify the grape diseases. We
also provide a practical study for dealing with the insuffi-
ciency and unbalance of dataset. The methods of data aug-
mentation techniques, early stopping mechanism and
dropout are used to improve the generalization ability of
model and reduce the risk of over-fitting. In addition, we have
proposed an effective multi-network integration method
which can be used to integrate more state-of-the-art CNN
models and can also be easily extended to other plant disease
identification tasks.
Declaration of Competing Interest
We declare that we have no financial and personal rela-
tionships with other people or organizations that can inap-
propriately influence our work, there is no professional or
other personal interest of any nature or kind in any product,
service and company that could be construed as influencing
the position presented in, or the review of, the manuscript
entitled ”Automatic Grape Leaf Diseases Identification via
UnitedModel Based on Multiple Convolutional Neural
Networks”.
Acknowledgements
This work was supported by the Public Welfare Industry (Agri-
culture) Research Projects Level-2 under Grant 201503116-04-
06; Postdoctoral Foundation of Heilongjiang Province under
Grant LBHZ15020; Harbin Applied Technology Research and
Development Program under Grant 2017RAQXJ096 and
National Key Application Research and Development Program
in China under Grant 2018YFD0300105-2.
R E F E R E N C E S
[1] Kole DK, Ghosh A, Mitra S. Detection of downy mildew
disease present in the grape leaves based on fuzzy set theory.
In: Advanced computing, networking and informatics. Berlin
(Germany): Springer; 2014. p. 377–84.
[2] Martinez A. Georgia plant disease loss estimates. link: http://
www.caes.uga. edu/Publications/displayHTML.cfm?pk_id=
7762. 2015.
[3] Srdjan S, Marko A, Andras A, Dubravko C, Darko S. Deep
neural networks based recognition of plant diseases by leaf
image classification. Comput Intell Neurosci 2016;2016:1–11.
[4] Wang G, Sun Y, Wang J. Automatic image-based plant disease
severity estimation using deep learning. Comput Intell
Neurosci 2017;2017:1–8.
[5] Xu G, Yang M, Wu Q. Sparse subspace clustering with low-
rank transformation. Neural Comput Appl 2017;2017:1–14.
[6] Singh V, Misra A. Detection of plant leaf diseases using image
segmentation and soft computing techniques. Inf Process
Agric 2017;4(1):41–9.
[7] Ma J, Du K, Zheng F, Zhang L, Gong Z, Sun Z. A recognition
method for cucumber diseases using leaf symptom images
based on deep convolutional neural network. Comput
Electron Agric 2018;154:18–24.
[8] Xu X, Liu Z, Wu Q. A novel K-nearest neighbor classification
algorithm based on maximum entropy. Int J Adv Comput
Technol 2013;5(5):966–73.
Fig. 8 – The trend of train accuracy, validation accuracy, train
loss and validation loss of the UnitedModel.
accuracy, respectively. In Fig. 8, we can observe that after a
few epochs they overlap almost completely, showing that
UnitedModel gets sufficiently trained.
Experimental results have shown that the proposed Uni-
tedModel integrating InceptionV3 and ResNet50 can achieve
promising results for the grape leaf diseases identification
task. However, since the training dataset used in our work
is composed of samples in a simple background scene, the
trained model cannot be applied for real-time diagnosis of
grape leaf diseases in the complex background. One direction
of future work is to create datasets under un-controled sce-
nario which can be used for training model to improve the
generalizability. Thus the proposed model can be used for
the development of mobile systems and devices. Another
direction of future work is to compress the model while keep-
ing the same performance. The proposed UnitedModel is a
combination of two popular models and is of huge number
the parameters. The training of the UnitedModel is great com-
putational cost. It is important to find out an effective pruning
mechanism for model compression to reduce computational
resources.
5.
Conclusions
In this work, we have developed an effective solution to auto-
matic grape diseases identification based on CNNs. We have
proposed the UnitedModel which is a united CNNs architec-
ture based on InceptionV3 and ResNet50 and can be used to
classify grape images into 4 classes, including 3 different
symptom images i.e., black rot, esca, isariopsis leaf spot,
and healthy images. UnitedModel takes advantage of the
combination of InceptionV3’s width and ResNet50’s depth
and thus can learn more representative features. The repre-
sentational ability of UnitedModel is strengthened by way of
high-level feature fusion, which makes it achieve the best
performance in the grape diseases identification task. The
experimental results demonstrate that our model can outper-
form the state-of-the-art basic single CNNs including VGG16,
InceptionV3, DenseNet121 and ResNet50. The proposed Uni-
tedModel achieves an average validation accuracy of 99.17%
and a test accuracy of 98.57% and thus can serve as a decision
Please cite this article as: M. Ji, L. Zhang and Q. Wu, Automatic grape leaf diseases identification via UnitedModel based on multiple convolu-
tional neural networks, Information Processing in Agriculture, https://doi.org/10.1016/j.inpa.2019.10.003
�











 2023年江西萍乡中考道德与法治真题及答案.doc
2023年江西萍乡中考道德与法治真题及答案.doc 2012年重庆南川中考生物真题及答案.doc
2012年重庆南川中考生物真题及答案.doc 2013年江西师范大学地理学综合及文艺理论基础考研真题.doc
2013年江西师范大学地理学综合及文艺理论基础考研真题.doc 2020年四川甘孜小升初语文真题及答案I卷.doc
2020年四川甘孜小升初语文真题及答案I卷.doc 2020年注册岩土工程师专业基础考试真题及答案.doc
2020年注册岩土工程师专业基础考试真题及答案.doc 2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc
2023-2024学年福建省厦门市九年级上学期数学月考试题及答案.doc 2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc
2021-2022学年辽宁省沈阳市大东区九年级上学期语文期末试题及答案.doc 2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc
2022-2023学年北京东城区初三第一学期物理期末试卷及答案.doc 2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc
2018上半年江西教师资格初中地理学科知识与教学能力真题及答案.doc 2012年河北国家公务员申论考试真题及答案-省级.doc
2012年河北国家公务员申论考试真题及答案-省级.doc 2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc
2020-2021学年江苏省扬州市江都区邵樊片九年级上学期数学第一次质量检测试题及答案.doc 2022下半年黑龙江教师资格证中学综合素质真题及答案.doc
2022下半年黑龙江教师资格证中学综合素质真题及答案.doc